Abstract
Histone deacetylases (HDACs) 1 and 2 share a high degree of homology and coexist within the same protein complexes. Despite their close association, each possesses unique functions. We show that the upregulation of HDAC2 in colorectal cancer occurred early at the polyp stage, was more robust and occurred more frequently than HDAC1. Similarly, while the expression of HDACs1 and 2 were increased in cervical dysplasia and invasive carcinoma, HDAC2 expression showed a clear demarcation of high-intensity staining at the transition region of dysplasia compared to HDAC1. Upon HDAC2 knockdown, cells displayed an increased number of cellular extensions reminiscent of cell differentiation. There was also an increase in apoptosis, associated with increased p21Cip1/WAF1 expression that was independent of p53. These results suggest that HDACs, especially HDAC2, are important enzymes involved in the early events of carcinogenesis, making them candidate markers for tumor progression and targets for cancer therapy.
Similar content being viewed by others
Introduction
Histone acetylases (HAT) and deacetylases (HDAC) are two opposing enzymes that regulate the transcriptional machinery by controlling the acetylation status of histones. The acetylation at ɛ-amino group of specific lysines in the N-terminus of histones neutralizes the positive charge on lysine. This is thought to reduce the affinity of histone complexes for DNA, enhancing the access of transcriptional factors to DNA. On the other hand, deacetylation results in a more condensed chromatin.1, 2, 3, 4, 5 There is clear evidence for the involvement of both HATs and HDACs in cell proliferation,6, 7, 8 differentiation9, 10, 11 and cell-cycle regulation.6, 12 Hence, the dysregulation of acetylation status in the cell is closely linked to cancer.13 Direct evidence for this comes from the study of the function of fusion proteins in leukemia.14, 15 PML-RARα and PLZF-RARα fusion proteins in acute promyelocytic leukemias recruit HDAC complexes, which repress genes that are necessary for differentiation. HDACs also function by interacting with tumor suppressor genes such as p53,16, 17, 18 Rb19, 20, 21 and BRCA1.22 The association of HDACs with these proteins results either in their recruitment to specific DNA promoter regions or in the deacetylation of the protein itself. For example, HDAC1 is recruited by the tumor suppressor Rb. The recruitment of HDAC1 is necessary for the repression of E2F target genes by Rb.1, 21 Alternatively, the deacetylation of nonhistone proteins such as p53 may also play a role in the control of cell cycle dynamics.17
Although there is clear evidence for the involvement of HDACs in the development of cancer, the specific roles of individual HDACs in the regulation of cell proliferation, apoptosis and cell cycle are unclear. The HDAC inhibitors used to demonstrate effects on cells are for the most part nonspecific for the different HDAC isoforms. There are at least 17 mammalian HDAC enzymes that have been classified into three groups based on homology to yeast HDAC enzymes and dependence on NAD+ for activity.5, 23, 24 Despite the increasing number of HDAC enzymes isolated from different species, the specialized functions of most HDACs are unknown. Also, the regulation of these enzymes during cell cycle and proliferation is largely unknown. Among the class I enzymes, HDAC1 and HDAC2 are shown to be highly homologous25, 26 and are found together in large protein complexes. These protein complexes are associated with classical transcriptional corepressors such as mSin327, 28, 29, 30 and NuRD.31, 32
In this study, we examined the expression of two closely related class I HDACs, namely HDAC1 and HDAC2 in matched samples of colon and cervical cancers. We show that HDAC2 is more consistently upregulated in colonic polyps and cancers than HDAC1. Both HDACs1 and 2 were upregulated in cervical dysplasias and carcinoma, but HDAC2 showed a stronger correlation with the dysplasia transition region compared to HDAC1. We also demonstrated the functional significance of HDAC2 upregulation by knocking down its expression using HDAC2-specific siRNA. HDAC2-knocked down cells displayed a differentiated morphology with reduction in cell density and increased apoptosis, associated with an increase in p21Cip1/WAF1 expression independent of p53. The data in this paper provide the first evidence for the different expression of these closely related HDAC enzymes in cancer and identifies histone deacetylases (HDACs), especially HDAC2, as candidate genes involved in the early events of carcinogenesis.
Results
HDAC1 and HDAC2 mRNA expression is increased in colorectal tumors
The expression of HDAC1 and HDAC2 mRNA was quantified in 16 colorectal cancers and matched normal mucosal samples using real-time quantitative RT-PCR. 18S mRNA was used to correct for minor variations in loading. The melting curve analysis and the sequencing results confirmed that the HDAC1 and HDAC2 RT-PCR products were specific (data not shown). The results are summarized in Figure 1 and Table 1. Only HDAC2 was significantly upregulated in matched colon tumor compared to normal mucosa (P<0.05).
HDAC2 was upregulated by at least two-fold in 9/16 tumor samples compared to matched normal mucosa. In contrast, only 6/16 showed a more than two-fold upregulation of HDAC1. Four of the 16 samples showed upregulation of HDAC2 by at least five-fold. HDAC2 was upregulated by more than eight-fold in three tumor samples. In contrast, HDAC1 was not upregulated by more than four-fold in all the tumor tissues examined. Five of the 16 sets of samples had concomitant polyps, which were also examined for both HDAC1 and HDAC2 mRNA expression. Three of the five polyps showed greater than two-fold upregulation of HDAC2 mRNA compared to normal mucosa. One of the polyps was upregulated by 12-fold. In contrast, only two of the five polyps showed more than two-fold upregulation of HDAC1.
HDAC1 and HDAC2 protein expression is increased in colorectal tumors
A tissue array was constructed from 45 matched samples of colorectal cancer and normal mucosa. The adequacy of the array was analyzed and all the tissue punches were scored by a pathologist (MST) according to the specifications stated in Materials and methods. In particular, the duplicate tissue array of 0.6-mm-diameter punches showed maximal (>95%) representativity of the intended tumor-normal samples. Furthermore, they allowed maximal correlation when comparing all the antibodies used, as immunohistochemistry was representative and successful in the vast majority of the punches. This is in keeping with our previous experience of TMA accurate representation on other neoplasms.33 Figure 2 illustrates a representative protein expression grading for both HDAC1 and HDAC2. Tables 2a and b show a summary of the grading intensities of the 45 matched pairs of samples expressed as percentages. The number of normal mucosal samples distributed in each of the grading intensities was similar for HDAC1 and HDAC2, suggesting that basal expression of the two proteins were similar in normal colonic mucosa. However, HDAC2 expression in tumors was higher compared to HDAC1. In all, 60% of the tumors were scored grade 3 for HDAC2 expression compared to 45% for HDAC1. Comparing normal mucosa and matched tumor samples, there was a 60% increase in the number of samples scoring 3 for HDAC2 compared to a 37% increase for HDAC1. Interestingly, 12.3% of the tumor samples had a score of 0 for HDAC1. None of the tumor samples were scored 0 for HDAC2 expression. The difference in HDAC1 and HDAC2 expression in tumors was more obvious when the differences in grading between tumor and matched normal mucosal samples were compared (Table 2c). There were significantly more tumor samples showing at least two-grade increase in intensity for HDAC2 staining compared to HDAC1 (P<0.05).
Colorectal TMA immunohistochemistry. Representative examples of the scoring performed on HDACs1 and 2 staining. Score 0 is defined as no positive staining in the nucleus; scores 1–3 represent increasing intensity of nuclear staining. The upper panel shows × 20 magnification and the lower panel shows × 600 magnification
p21Cip1/WAF1 expression is inversely correlated with HDAC2 in colorectal cancer
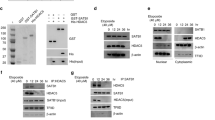
p21Cip1/WAF1 was expressed at the upper two-thirds of a normal colon crypt but was not detected at the base of the crypt (Figure 3a). The expression of HDAC2, however, showed the opposite pattern, with expression predominantly at the base of the crypt (Figure 3b). p21Cip1/WAF1 nuclear expression was detected in 88% of the normal samples but only in 22% of the tumor samples. This distribution is consistent with the inverse correlation between HDAC2 and p21Cip1/WAF1 expression since HDAC2 expression was higher in tumor samples. When a comparison was made between tumor samples and their respective matched normal, 86% of the tumor samples showing upregulation of HDAC2 had concomitant downregulation of p21Cip1/WAF1.
Expression of p21Cip1/Waf1 is inversely correlated to HDAC2 in the normal colon and cervix. High expression of p21Cip1/Waf1 was observed at the upper two-thirds of the crypt ( → ) ( × 100) (a), whereas HDAC2 expression was limited to the base of the crypt ( → ) ( × 200) (b). p21Cip1/Waf1 expression was limited to the middle zone (MZ) of the cervical epithelium. No staining was seen in the basal proliferative layer ( → ) ( × 200) (c)
HDAC1 and HDAC2 protein expression is correlated with cervical dysplasia
Figure 4 shows representative samples of different stages of cervical intraepithelial neoplasia (CIN) stained with specific HDAC1 and HDAC2 antibodies. Serial sections were used for immunohistochemistry. Both HDACs1 and 2 showed similar pattern of staining in the normal epithelium as well as invasive carcinoma. They were strongly expressed in the proliferating basal cells of the normal epithelium (Figures 4a and e). Even though both HDACs1 and 2 expression was correlated with the degree of dysplasia, HDAC2 staining showed a clearer demarcation of upregulation in the dysplasia transition region compared to HDAC1 (Figures 4b and f). Moreover, less number of cells was positively stained with HDAC1 compared to HDAC2 in the dysplastic region. Invasive carcinoma was strongly stained for both HDAC1 and 2 (Figures 4d and h). Similar to the observation in normal colon tissues, p21Cip1/WAF1 expression in the normal cervical tissue was also inversely correlated to the expression of HDACs1 and 2. p21Cip1/WAF1 expression was absent in the basal layer of proliferating epithelial cells where HDACs1 and 2 were strongly expressed, and positive in the middle zone where HDACs1 and 2 were weakly expressed (Figure 3c).
HDAC2 knockdown using siRNA in HeLa cells is associated with an increase in p21Cip1/WAF1 expression
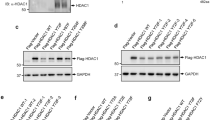
The function of HDAC2 in cell growth was examined by knocking down HDAC2 expression in HeLa cells. A HDAC2 sequence-specific small interfering RNA (siRNA) was designed and transiently transfected into the cells. HeLa cells were chosen because they were readily transfectable and produced consistent knockdown of at least 50% when compared to scrambled siRNA-transfected cells. Figure 5 is a Western blot showing HDAC2 knockdown in HeLa cells transfected with 4.8 μg HDAC2 siRNA (HD2) compared to 4.8 μg scrambled (scr) siRNA-transfected cells at 72 h post-transfection. The samples were also probed for HDAC1, p21Cip1/WAF1, p53 and GAPDH. There was no knockdown of HDAC1 mRNA and protein, even though the protein sequence is highly homologous to HDAC2. The GAPDH protein was used as a control for protein loading. The knockdown of HDAC2 in the cells was evident at 24 h post-transfection. At 48 h after transfection, HDAC2 knockdown caused a 1.8-fold increase in p21Cip1/WAF1 mRNA and a four-fold increase in protein. The induction of mRNA and protein were maintained for at least 72 h. HDAC2 knockdown also resulted in a 1.7-fold increase in p53 mRNA 48 h after transfection, but protein levels remain unchanged. The results are representative of four independent experiments.
HDAC2 knockdown results in a change in cell density and morphology
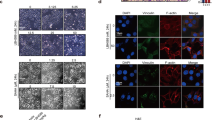
Treatment of cells with HDAC2-specific siRNA resulted in changes in cell density and morphology. Figure 6a is a representative field of HeLa cells 96 h after transfection with 4.8 μg of HDAC2-specific siRNA. The cells were sparse compared to cells treated with scrambled siRNA, which were confluent at this time (Figures 6a and b). Cells in the HDAC2 siRNA group showed morphological characteristics of apoptosis as characterized by their being rounded, shrunken and highly refractile. Interestingly, the remaining cells were flatter and had more extensions compared to the scrambled treated cells. This morphology is reminiscent of a differentiated phenotype. For comparison, the morphology of cells treated with butyrate, a known differentiating agent, is shown in Figure 6c. HeLa cells treated with butyrate were flat and had numerous extensions.
HDAC2 knockdown increases apoptosis in HeLa cells
The functional significance of HDAC2 knockdown was examined by determining its effect on apoptosis. The cells were stained with Annexin V and propidium iodide (PI) at 72 h post-transfection. Figure 7a is a representative dot plot analysis of PI against Annexin V staining. Apoptotic cells are represented in quadrant 2 (indicated by the numerical percentage). Figure 7b summarizes data from four independent experiments and shows a significant increase in the percentage of apoptotic cells in the HDAC2 knockdown cells compared to the scrambled population after 72 h (18.5 versus 8.2%, P<0.05). The increase in apoptosis was also evident in Figure 7c, which is a graphical plot of the number of cells against PI staining (4.1% against 13.0%). The sub-G1 fraction (M1) represents the apoptotic cell population. There was no significant difference in the distribution of cells in the other cell-cycle phases. There was also no apparent cell cycle arrest at either G1/S or G2M.
Apoptosis assay on HDAC2-knocked down HeLa cells. (a) Annexin V assay dot plot; the percentage of population undergoing early apoptotic events is indicated in the lower right quadrant. (b) Percentage of early apoptotic cells in four independent experiments (72 h post-transfection). *P<0.01. (c) Cell-cycle analysis using PI staining
Discussion
The results from this study are the first to compare HDACs1 and 2 regulation in human colon and cervical cancer. HDACs1 and 2 mRNA and protein levels were upregulated in colorectal cancer. However, the upregulation of HDAC2 was more robust and observed more frequently compared to HDAC1. HDAC2 mRNA expression was upregulated by more than two-fold in 9/16 tumors, compared to 6/16 for HDAC1. Similarly, 61.1% of tumors showed a two or three grade difference in intensity of HDAC2 staining, compared to 40.3% of tumors for HDAC1. Thus, there were significantly more tumor samples showing at least two-grade increase in intensity for HDAC2 staining compared to HDAC1 (P<0.05). A significantly greater number of samples showed an increase in HDAC2 staining compared to HDAC1 (P<0.05). The upregulation of HDAC2 was observed in colonic polyps, suggesting that the change occurs early in the carcinogenic process. The transition from polyp to carcinoma was also accompanied by a further increase in HDAC2 mRNA expression in all five polyps examined. In contrast, the upregulation of HDAC1 was not as robust in polyps. Only two of the five polyps showed an increase in expression in HDAC1 compared to matched normal mucosa. Further, only two of the five polyps showed a change in HDAC1 expression when compared to matched colonic tumors. This suggests that HDACs1 and 2, although closely related, sharing 75% identity in DNA sequence and 85% identity in protein sequence,25, 26 and often found together in protein complexes, may be regulated differently in colorectal polyps and colonic cancer. The different regulation of HDAC1 and 2 in cancer is consistent with the proposal that they have distinct functions in the cell. Lagger et al.,34 using HDAC1-deficient mouse embryonic stem cells, showed that HDAC1 possessed a unique function that could not be compensated by HDAC2. A separate study showed that HDAC1 and 2 have distinct functions in senescence.35 The investigators isolated a senescence-specific form of HDAC2 that did not contain HDAC1, a form not seen in younger cells.
The upregulation of both HDACs1 and 2 was also observed in cervical dysplasia and invasive carcinoma. Interestingly, both HDACs1 and 2 were expressed in the basal layer of normal cervical tissue. No expression was detected in the upper layers of the epithelium, comprising the more mature and differentiated cells, suggesting that the expression of both isoforms was correlated to cell proliferation. The pattern of upregulation of HDAC1 and 2 was similar in cervical dysplasia and invasive carcinoma. However, HDAC2 showed a more distinct demarcation of higher staining intensity at the dysplasia transition region. Further, there appeared to be more cells staining positive for HDAC2 than HDAC1 in cervical dysplasia, consistent with the observation in colorectal polyps and cancer, where HDAC2 was more strongly upregulated than HDAC1.
The study also highlights the variations in HDAC1 and 2 expression in normal tissue and cancer. This was particularly apparent in the immunohistochemistry study using the tissue microarray (TMA). The normal mucosal staining intensities for HDAC1 ranged from grades 0 to 3, while HDAC2 from grades 0 to 2. The tumor staining intensities were also widely distributed, although they were skewed towards the higher staining intensities. It is only when the relative staining intensities of matched samples were compared that the increase in expression in cancer for both HDAC1 and 2 became apparent. It is therefore not surprising that the differences in HDAC expression in cancer was not detected in an analysis using nonmatched samples.23
Our results suggest that an increase in HDAC activity, especially that of HDAC2, facilitates the development of tumors. This hypothesis is supported by our results showing that the knockdown of HDAC2 in cells results in increased apoptosis. Consistent with this, recent results from Zhu et al.,36 show that overexpression of HDAC2 protects cells against apoptosis. The protection of cells against apoptosis by HDAC2 may be an important factor facilitating the development of tumors. Taken together, these results suggest that the upregulation of HDAC2 is a cause, rather than a consequence of carcinogenesis. Consistent with its role in carcinogenesis, HDAC2 expression was upregulated early in this process, at the colonic polyp and cervical dysplastic stages. This proposal is consistent with studies showing that the inhibition of HDAC activity using a variety of inhibitors reverses the cancer phenotype by causing cell differentiation,13, 24, 37, 38, 39 apoptosis40, 41, 42, 43, 44 and cell-cycle arrest.45, 46 However, since the inhibitors used were not specific for any particular HDAC isoform, it has not been possible to determine the functional significance of these isoforms in cancer development. Our results indicate that the specific HDAC isoform HDAC2 plays an important role in cancer development independent of HDAC1, since the reduction in HDAC2 expression occurred without a decrease in HDAC1 expression (Figure 5).
The increase in p21Cip1/WAF1 mRNA and protein after HDAC2 knockdown suggests that HDAC2 may be involved in the regulation of p21Cip1/WAF1 at the transcriptional level. This hypothesis is further supported by our immunohistochemistry data showing an inverse correlation between HDAC2 and p21Cip1/WAF1 expression in both normal colon and cervical tissue. The inverse relationship was also observed in colon tumor samples. Interestingly, the expression of p21Cip1/WAF1 in tumor cells overexpressing HDAC2 was not only reduced but also aberrantly localized to the cytoplasm instead of the nucleus. Studies have shown that p21Cip1/WAF1 cytoplasmic expression may render its growth inhibitory function ineffective.47, 48 Hence, we postulate that HDAC2 can regulate both the expression and localization of p21Cip1/WAF1 in the cell.
As p53 is a known regulator of p21Cip1/WAF1 expression, we determined the levels of p53 mRNA and protein in the cell after HDAC2 knockdown. Our results indicate that the change in p21Cip1/WAF1 mRNA and protein occurred without a change in p53 protein levels, suggesting that HDAC2 knockdown increases p21Cip1/WAF1 independent of p53. Our results also suggest that the increase in apoptosis, consequent to HDAC2 knockdown, is independent of p53. Hence, we suggest that apoptosis induced by a knockdown of HDAC2 involves p21 but not p53. The mechanism by which p21Cip1/WAF1 may cause apoptosis is currently unclear. There are reports that suggest that p21Cip1/WAF1 inhibits apoptosis. Hence, downregulation of p21Cip1/WAF1 renders cells more sensitive to interferon-, cytotoxic- and gamma irradiation-induced apoptosis.49, 50, 51, 52 p21Cip1/WAF1has also been shown to inhibit Fas-mediated apoptosis by forming a complex with procaspase-3.53 However, p21Cip1/WAF1 has been shown to induce apoptosis or even found to be indispensable for apoptosis.54 Increasing p21Cip1/WAF1 expression has been shown to induce apoptosis in glioma, hepatoma, cervical, ovarian and mammary tumor cells.55, 56 It is possible that the level of p21Cip1/WAF1 in the cell is an important determining factor as to whether the cell differentiates or undergoes apoptosis. For example, lower levels of p21Cip1/WAF1 may cause the cell to differentiate and higher levels may direct the cell to an apoptotic pathway. The relationship between the upregulation of p21Cip1/WAF1 in our study after HDAC2 knockdown and apoptosis is currently unknown. It is possible that the extremely high levels of p21Cip1/WAF1 induced (up to six-fold) in the cell may direct the cell to an apoptotic pathway. However, it is also possible, from the morphological changes observed, that cells which may not be expressing such high levels of p21Cip1/WAF1 may be induced to differentiate. Further experiments will be required to address this possibility. It would be interesting to compare the relative effects of HDAC1 and 2 knockdowns on apoptosis and differentiation.
In conclusion, the study has shown, for the first time, that the expression of HDACs1 and 2 is increased in colorectal polyp and cancer, and in cervical dysplasia and carcinoma. The upregulation of HDAC2 expression was more robust and occurred more frequently in colorectal tumors. The upregulation occurred at an early phase in colorectal cancer development, at the polyp stage. We have also demonstrated that HDAC2 knockdown induces apoptosis possibly through an upregulation of p21Cip1/WAF1, but independent of p53. HDAC2 may also play a role in the differentiation status of a cell. Taken together, the study suggests that HDACs, especially HDAC2, are important markers for cancer progression and candidate targets for drug therapy.
Materials and Methods
Fresh tissue samples and RNA isolation
Anonymized samples of fresh tumor and matched normal mucosa were obtained from 16 human colorectal cancers surgically resected between June 1997 to August 2000 at National University Hospital and Tan Tock Seng Hospital, Singapore. Five of these sets of samples had concomitant polyps. All samples were frozen and stored in liquid nitrogen until ribonucleic acid (RNA) extraction. Histopathology of the samples was verified by a pathologist. Total RNA was extracted from the tissues by the guanidinium thiocyanate method.57 RNA was quantified photometrically at 260 nm.
Quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR)
Real-time RT-PCR was carried out using the LightCycler System instrument (Roche, Mannheim, Germany). Amplified products were detected by measuring the binding of the fluorescence dye SYBR Green I to double-stranded DNA (SYBR Green I RNA amplification kit, Roche). Primers were designed and optimized for maximum efficiency and specificity according to the manufacturer's specifications. The primer sequences used were as follows: HDAC2 forward: 5′-GTTGCTCGATGTTGGAC-3′, reverse: 5′-CCAGGTGCATGAGGTA-3′; HDAC1 forward: 5′-AACTGGGGACCTACGG-3′, reverse: 5′-ACTTGGCGTGTCCTT-3′;18S ribosomal RNA forward: 5′-GTAACCCGTTGAACCCCATT-3′, reverse: 5′-CCATCCAATCGGTAGTAGCG-3′; GAPDH forward: 5′-AGCAATGCCTCCTGCACCACCAAC-3′; reverse: 5′-CCGGAGGGGCCATCCACAGTCT-3′; p21Cip1/WAF1 forward: 5′-ATTAGCAGCGGAACAAGGAGTCAGACAT-3′, reverse: 5′-CTGTGAAAGACACAGAACAGTACAGGGT-3′ and p53 forward: 5′-CACTAAGCGAGCACTG-3′, reverse: 5′-GGAGGTAGACTGACCC-3′. The PCR reaction components comprised 2 μl of RT-PCR reaction mix (dNTPs, MgCl2, SYBR Green), and 0.2 μl of RT-PCR enzyme mix, a final concentration of 6 mM MgCl2, and 0.4 μl of each primer (10 μM), in a total volume of 10 μl. The PCR reaction was started with an initial reverse transcription at 55°C for 10 min, followed by 30 s at 95°C and 45 cycles of amplification (0 s at 95°C, 10 s at 60°C (62°C for p21Cip1/WAF1) and 9 s (13 s for p21Cip1/WAF1) at 72°C) for all primer sets. At the end of each cycle, the fluorescence emitted by SYBR Green was measured. After completion of the amplification process, samples were subjected to a temperature ramp with continuous fluorescence monitoring for melting curve analysis to test for product specificity. The products were analyzed by electrophoresis on a 2% agarose gel and verified by sequencing. A serial dilution of standards was individually constructed for each set of primers to determine the log-linear range and the efficiency of the reaction using the LightCycler Data Analysis Software. Relative quantification of HDAC1 and 2 in normal, polyp and tumor samples was carried out after normalization with 18S, an internal control. For cell lines, GAPDH was used as an internal control. Calculations were performed according to the manufacturer's specifications.
Tissue microarray
A total of 90 colorectal samples (45 sets of tumor and paired normal mucosa) were included in this study. These were random cases from the files of the Department of Pathology, National University Hospital of Singapore, with no selection bias regarding gender, age, clinical presentation or tumor staging. The materials were fully anonymized prior to the inclusion in the study. TMAs were constructed as before,33 including negative and positive controls in the array to assess the adequacy of the staining. After a morphologically representative area of tumor was annotated by the pathologist, tissue cylinders with a 0.6 mm diameter were punched from the donor tissue block and deposited into a recipient block using a tissue arraying instrument (Beecher Instruments®, Silver Spring, MD, USA). A section was stained with hematoxylin and eosin (H&E) for histological confirmation of the arrayed tissues. Scoring of HDAC1 and 2 expressions in the TMA format was based on the intensity of the staining, with 0 score if no staining was detected and 1 to 3 representing low, moderate or intense expression (see Figure 2 for an illustration of this approach). The presence of p21Cip1/WAF1 was based on positive nuclear staining.
Cervical tissue blocks
Nine blocks of tissue from uterine cervix showing CIN of different grades were selected for this study. These blocks were randomly selected from the files of Department of Pathology, National University of Singapore and were fully anonymized prior to the inclusion in the study.
Immunohistochemistry
Immunohistochemistry was performed using a standard indirect immunoperoxidase method. Sections from formalin-fixed, paraffin-embedded tissues were deparaffinized, treated with 3% hydrogen peroxide in Tris-buffered saline and pretreated at 96°C for 30 min in 10 μmol/l citrate buffer (pH 6.0). For p21Cip1/WAF1 staining, the sections were treated with Antigen Unmasking Solution (Vector Laboratories, California, USA). Staining was carried out by an avidin-biotinylated horseradish peroxidase complex method (DAKO, Glostrup, Denmark) using a rabbit polyclonal antibody against HDAC2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), a rabbit polyclonal antibody against HDAC1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), a mouse monoclonal antibody against p21Cip1/WAF1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The dilutions used for the HDAC1, HDAC2 and p21Cip1/WAF1 antibodies were 1 : 100, 1 : 500 and 1 : 100 respectively. HDAC1 and 2 antibodies have no crossreactivity with each other as unique regions were used to design each of the antigen. Hematoxylin was used as counterstain.
Cell culture and reagents
The human cervical carcinoma HeLa cells were purchased from ATCC (Bethesda, MD, USA) and grown in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA). Cells were trypsinized, seeded at 1.5 × 105 cells/well in a six-well plate, and allowed to grow overnight at 37°C in a humidified incubator at 5% CO2. At 24 h after plating, the cells were transfected with 1, 2 and 4.8 μg of HDAC2-specific siRNA and scrambled siRNA in serum-free and antibiotic-free RPMI 1640 medium. Transfection was carried out using GeneSilencer™ siRNA transfection reagent (Gene Therapy Systems Inc., San Diego, CA, USA) according to the manufacturer's specifications. At 4 h after transfection, an equal volume of medium with 20% FBS supplement was added to each well. The cells were washed with fresh medium 24 and 48 h after transfection, and maintained in a 10% FBS-supplemented medium. The cells were harvested at specified time points for protein and RNA extraction, assayed for apoptosis, Western analysis and quantitative real-time RT-PCR. As a positive control for cell morphology changes, HeLa cells were treated with 5 mM sodium butyrate for 72 h.
HDAC2 siRNA
The HDAC2-specific siRNA duplex was designed using the OligoEngine™ software (www.oligoengine.com) according to its set of algorithms. The siRNA was synthesized by Dharmacon Inc. (Lafayette, CO, USA) and delivered in 2′-deprotected and desalted form. An siRNA duplex was designed against the HDAC2 gene to recognize the sequence 5′-AATCCGCATGACCCATAACTT-3′. Scrambled siRNA was also purchased through OligoEngine™ (Cat. No. D1200-20) and used as a control.
Annexin assay and PI staining
Annexin-V-FLUOS Staining Kit (Roche Diagnostics GmbH, Germany) was used to quantify the level of apoptosis in the samples, according to the manufacturer's specifications. Briefly, cells were trypsinized, centrifuged and resuspended in 100 μl of Annexin-V-FLUOS labeling solution. After a 15-min incubation, the cells were analyzed using FACS Vantage SE Flow Cytometry system (Becton Dickinson, NJ, USA). A separate aliquot of cells was also stained with 50 μg/ml PI in the presence of 0.6% NP-40 and 0.2 mg/ml RNAse A, for cell-cycle analysis.
Western blot analysis
Total protein extraction was carried out using lysis buffer (6 M urea, 1% 2-mercaptoethanol, 50 mM Tris buffer (pH 7.4), 1% SDS in phosphate-buffered saline (pH 7.4)). Total protein (30 μg) was subjected to SDS-PAGE on a 4% stacking gel and 12% resolving gel for the analysis of HDAC1, HDAC2, p21Cip1/WAF1, p53 and GAPDH expressions. Proteins were transferred to a nitrocellulose membrane (Hybond C-Extra, Amersham Biosciences, UK) and blocked overnight in PBS solution containing 0.1% Tween-20 and 5% (w/v) nonfat milk powder. The membrane was then incubated with anti-HDAC1, anti-HDAC2, anti-p53, anti- p21Cip1/WAF1 (Santa Cruz Biotechnologies Inc., Santa Cruz, CA, USA) and anti-GAPDH (Chemicon International Inc., Ternecula, CA, USA) for 1 h and subsequently with an anti-rabbit or an anti-mouse secondary antibody. SuperSignal® West Dura (Pierce Biotechnology Inc., Rockford, IL, USA) chemiluminescent substrate was used to detect the bound antibodies. The membrane was then exposed to Kodak BioMax film (Eastman Kodak, Rochester, NY, USA). The intensity of the Western blot bands was quantified using Typhoon 8600 (Molecular Dynamics, division of Amersham Pharmacia Biotech, Sunnyvale, CA, USA).
Statistical test
Paired samples t-test and χ2 test with Yates' continuity correction were performed using GraphPad Prism Version 4.0. A two-tailed P-value of less than 0.05 was considered statistically significant.
Abbreviations
- HDAC:
-
histone deacetylase
- HAT:
-
histone acetylase
- siRNA:
-
small interfering RNA
- TMA:
-
tissue microarray
References
Wade PA (2001) Transcriptional control at regulatory checkpoints by histone deacetylases: molecular connections between cancer and chromatin. Hum. Mol. Genet. 10: 693–698
Strahl BD and Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45
Ito K, Barnes PJ and Adcock IM (2000) Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol. 20: 6891–6903
Forsberg EC and Bresnick EH (2001) Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23: 820–830
Cress WD and Seto E (2000) Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 184: 1–16
Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A and Levrero M (1997) p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16: 369–383
Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM and Eckner R (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93: 361–372
Walker SS, Shen WC, Reese JC, Apone LM and Green MR (1997) Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90: 607–614
Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y and Levrero M (1997) Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1: 35–45
Eckner R, Yao TP, Oldread E and Livingston DM (1996) Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10: 2478–2490
Kawasaki H, Eckner R, Yao TP, Taira K, Chiu R, Livingston DM and Yokoyama KK (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393: 284–289
Suzuki-Yagawa Y, Guermah M and Roeder RG (1997) The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol. Cell. Biol. 17: 3284–3294
Marks P, Rifkind RA, Richon VM, Breslow R, Miller T and Kelly WK (2001) Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1: 194–202
Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Lazar MA, Minucci S and Pelicci PG (1998) Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature 391: 815–818
Lin RJ, Nagy L, Inoue S, Shao W, Miller Jr WH and Evans RM (1998) Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391: 811–814
Luo J, Su F, Chen D, Shiloh A and Gu W (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381
Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS and Wu CW (2000) Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275: 20436–20443
Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ and George DL (1999) Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13: 2490–2501
Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ and Kouzarides T (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391: 597–601
Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D and Harel-Bellan A (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391: 601–605
Luo RX, Postigo AA and Dean DC. (1998) Rb interacts with histone deacetylase to repress transcription. Cell 92: 463–473
Yarden RI and Brody LC (1999) BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96: 4983–4988
De Ruijter AJ, Van Gennip AH, Caron HN, Kemp S and Van Kuilenburg AB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370 (Part 3): 737–749
Johnstone RW (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 1: 287–299
Yang WM, Inouye C, Zeng Y, Bearss D and Seto E (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 93: 12845–12850
Taunton J, Hassig CA and Schreiber SL (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411
Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P and Reinberg D (1997) Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89: 357–364
Laherty CD, Yang WM, Sun JM, Davie JR, Seto E and Eisenman RN (1997) Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89: 349–356
Knoepfler PS and Eisenman RN (1999) Sin meets NuRD and other tails of repression. Cell 99: 447–450
Hassig CA, Fleischer TC, Billin AN, Schreiber SL and Ayer DE (1997) Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89: 341–347
Xue Y, Wong J, Moreno GT, Young MK, Cote J and Wang W (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2: 851–861
Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A and Reinberg D (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13: 1924–1935
Zhang D, Salto-Tellez M, Putti TC, Do E and Koay ES (2003) Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod. Pathol. 16: 79–84
Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T and Seiser C (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21: 2672–2681
Wagner M, Brosch G, Zwerschke W, Seto E, Loidl P and Jansen-Durr P (2001) Histone deacetylases in replicative senescence: evidence for a senescence-specific form of HDAC-2. FEBS Lett. 499: 101–106
Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP and Gottlicher M (2004) Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell 5: 455–463
Marks PA, Richon VM and Rifkind RA (2000) Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 92: 1210–1216
Kramer OH, Gottlicher M and Heinzel T (2001) Histone deacetylase as a therapeutic target. Trends Endocrinol. Metab. 12: 294–300
Kosugi H, Towatari M, Hatano S, Kitamura K, Kiyoi H, Kinoshita T, Tanimoto M, Murate T, Kawashima K, Saito H and Naoe T (1999) Histone deacetylase inhibitors are the potent inducer/enhancer of differentiation in acute myeloid leukemia: a new approach to anti-leukemia therapy. Leukemia 13: 1316–1324
Weidle UH and Grossmann A (2000) Inhibition of histone deacetylases: a new strategy to target epigenetic modifications for anticancer treatment. Anticancer Res. 20: 1471–1485
Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, Kofler R, Smyth MJ and Johnstone RW (2001) The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc. Natl. Acad. Sci. USA 98: 10833–10838
Vrana JA, Decker RH, Johnson CR, Wang Z, Jarvis WD, Richon VM, Ehinger M, Fisher PB and Grant S (1999) Induction of apoptosis in U937 human leukemia cells by suberoylanilide hydroxamic acid (SAHA) proceeds through pathways that are regulated by Bcl-2/Bcl-XL, c-Jun, and p21CIP1, but independent of p53. Oncogene 18: 7016–7025
Kwon SH, Ahn SH, Kim YK, Bae GU, Yoon JW, Hong S, Lee HY, Lee YW, Lee HW and Han JW (2002) Apicidin, a histone deacetylase inhibitor, induces apoptosis and Fas/Fas ligand expression in human acute promyelocytic leukemia cells. J. Biol. Chem. 277: 2073–2080
Glick RD, Swendeman SL, Coffey DC, Rifkind RA, Marks PA, Richon VM and La Quaglia MP (1999) Hybrid polar histone deacetylase inhibitor induces apoptosis and CD95/CD95 ligand expression in human neuroblastoma. Cancer Res. 59: 4392–4399
Finzer P, Kuntzen C, Soto U, zur Hausen H and Rosl F (2001) Inhibitors of histone deacetylase arrest cell cycle and induce apoptosis in cervical carcinoma cells circumventing human papillomavirus oncogene expression. Oncogene 20: 4768–4776
Sandor V, Senderowicz A, Mertins S, Sackett D, Sausville E, Blagosklonny MV and Bates SE (2000) P21-dependent g(1)arrest with downregulation of cyclin D1 and upregulation of cyclin E by the histone deacetylase inhibitor FR901228. Br. J. Cancer 83: 817–825
Coqueret O (2003) New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 13: 65–70
Heliez C, Baricault L, Barboule N and Valette A (2003) Paclitaxel increases p21 synthesis and accumulation of its AKT-phosphorylated form in the cytoplasm of cancer cells. Oncogene 22: 3260–3268
Detjen KM, Murphy D, Welzel M, Farwig K, Wiedenmann B and Rosewicz S (2003) Downregulation of p21(waf/cip-1) mediates apoptosis of human hepatocellular carcinoma cells in response to interferon-gamma. Exp. Cell Res. 282: 78–89
Tian H, Wittmack EK and Jorgensen TJ (2000) p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 60: 679–684
Mahyar-Roemer M and Roemer K (2001) p21 Waf1/Cip1 can protect human colon carcinoma cells against p53-dependent and p53-independent apoptosis induced by natural chemopreventive and therapeutic agents. Oncogene 20: 3387–3398
Han Z, Wei W, Dunaway S, Darnowski JW, Calabresi P, Sedivy J, Hendrickson EA, Balan KV, Pantazis P and Wyche JH (2002) Role of p21 in apoptosis and senescence of human colon cancer cells treated with camptothecin. J. Biol. Chem. 277: 17154–17160
Suzuki A, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y and Akahane K (2000) Procaspase 3/p21 complex formation to resist fas-mediated cell death is initiated as a result of the phosphorylation of p21 by protein kinase A. Cell Death Differ. 7: 721–728
Chopin V, Toillon RA, Jouy N and Le Bourhis X (2004) P21(WAF1/CIP1) is dispensable for G1 arrest, but indispensable for apoptosis induced by sodium butyrate in MCF-7 breast cancer cells. Oncogene 23: 21–29
Tsao YP, Huang SJ, Chang JL, Hsieh JT, Pong RC and Chen SL (1999) Adenovirus-mediated p21((WAF1/SDII/CIP1)) gene transfer induces apoptosis of human cervical cancer cell lines. J. Virol. 73: 4983–4990
Shibata MA, Yoshidome K, Shibata E, Jorcyk CL and Green JE (2001) Suppression of mammary carcinoma growth in vitro and in vivo by inducible expression of the Cdk inhibitor p21. Cancer Gene Ther. 8: 23–35
MacDonald RJ, Swift GH, Przybyla AE and Chirgwin JM (1987) Isolation of RNA using guanidinium salts. Methods Enzymol. 152: 219–227
Acknowledgements
This work was supported by grants from Academic Research Fund (R-185-000-039-112) and National Medical Research Council (R-185-000-041-213) of Singapore. The Tissue Microarray work was supported by the Singapore Cancer Syndicate grant (SCS-MN5).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by M Piacentini
These authors contributed equally to this work
Rights and permissions
About this article
Cite this article
Huang, B., Laban, M., Leung, CW. et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ 12, 395–404 (2005). https://doi.org/10.1038/sj.cdd.4401567
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401567
Keywords
This article is cited by
-
UBE4B interacts with the ITCH E3 ubiquitin ligase to induce Ku70 and c-FLIPL polyubiquitination and enhanced neuroblastoma apoptosis
Cell Death & Disease (2023)
-
Antineoplastic activity of plant-derived compounds mediated through inhibition of histone deacetylase: a review
Amino Acids (2023)
-
In silico profiling of histone deacetylase inhibitory activity of compounds isolated from Cajanus cajan
Beni-Suef University Journal of Basic and Applied Sciences (2022)
-
Roles of HDAC2, eIF5, and eIF6 in Lung Cancer Tumorigenesis
Current Medical Science (2021)
-
Targeted therapy for mTORC1-driven tumours through HDAC inhibition by exploiting innate vulnerability of mTORC1 hyper-activation
British Journal of Cancer (2020)