Abstract
Growth hormone is used to increase height in short children who are not deficient in growth hormone, but its efficacy varies largely across individuals. The genetic factors responsible for this variation are entirely unknown. In two cohorts of short children treated with growth hormone, we found that an isoform of the growth hormone receptor gene that lacks exon 3 (d3-GHR) was associated with 1.7 to 2 times more growth acceleration induced by growth hormone than the full-length isoform (P < 0.0001). In transfection experiments, the transduction of growth hormone signaling through d3-GHR homo- or heterodimers was ∼30% higher than through full-length GHR homodimers (P < 0.0001). One-half of Europeans are hetero- or homozygous with respect to the allele encoding the d3-GHR isoform, which is dominant over the full-length isoform. These observations suggest that the polymorphism in exon 3 of GHR is important in growth hormone pharmacogenetics.
Similar content being viewed by others
Main
Response to growth hormone therapy can be considered a continuous quantitative trait that has measurable phenotypic variation resulting from both genetic and nongenetic effects. Response to growth hormone therapy is variable in short children who are not deficient in growth hormone1,2,3. Age, body mass and growth hormone dose have a role in this individual variability, but the genetic factors influencing response to growth hormone ('growth hormone pharmacogenetics') are still unknown. Candidate genes, however, can easily be identified from the molecular network by which target tissues respond to growth hormone. The first step of growth hormone action is the binding of growth hormone to the growth hormone receptor (GHR), followed by the activation of the JAK-STAT pathway and subsequent increase in expression of insulin-like growth factor 1 (IGF1) and other growth hormone–dependent genes. GHR consists of an extracellular domain of 246 amino acids, a single transmembrane domain and a cytoplasmic domain. The human gene GHR contains nine coding exons. Exons 3–7 encode the extracellular domain. There are two isoforms of GHR in humans, generated by retention or exclusion of exon 3 during splicing: a full-length isoform and an isoform that lacks exon 3 (d3-GHR). The generation of two transcripts that differ by the skipping of a coding exon results from homologous recombination, which mimics alternative splicing between the two retroviral sequences that flank the skipped exon4. The allele encoding d3-GHR is therefore specific to humans. The importance of the region encoded by exon 3 is unknown. The region is conserved in GHR proteins in mammalian species but is absent in the prolactin receptor5. This pattern of evolutionary conservation suggests that the loss or retention of exon 3 could affect receptor expression or function, specifically by affecting binding of human growth hormone, receptor processing, transport, stability, binding to other ligands, dimerization of GHR monomers or signal transduction.
The 22 residues encoded by exon 3 could not be modeled from the crystal structure of the complex between growth hormone and the extracellular domain of GHR6, but the peptide is located away from binding interfaces. The intramolecular disulfide bond pattern of the extracellular domain does not seem to be affected by the loss of exon 3, and so the global folding of the extracellular domain is supposed not to be altered. Exclusion of exon 3 results in the loss of one potential glycosylation site and the substitution A6D at the end of exon 2. This latter modification involves a highly conserved amino acid and leads to a change in charge, size and hydrophobicity of the receptor domain.
The binding of growth hormone to the two GHR isoforms has been studied in various experiments. When transiently expressed in COS-7 cells, full-length GHR and d3-GHR have comparable growth hormone–binding properties7. In another study, mRNAs were transcribed from cDNA encoding human full-length GHR and d3-GHR and microinjected into Xenopus laevis oocytes8. d3-GHR was as efficient as full-length GHR in binding and internalizing growth hormone in this study, but both isoforms also bound human placental lactogen and ovine prolactin, calling into question the specificity of the binding assay. In other experiments using soluble binding proteins, monomeric d3-GHR binding protein and full-length GHR binding protein showed no difference in binding to recombinant human growth hormone9. In addition, recent clinical data indicate that a single allele encoding d3-GHR is functional in humans10. To our knowledge, however, there has been no comparative evaluation of the efficacy of the two GHR isoforms in transducing the signals generated by growth hormone binding.
After the dimerization of two transmembrane chains to form functional GHR, homozygous individuals have either full-length GHR or d3-GHR homodimers on their cell surfaces, whereas heterozygotes have full-length GHR homodimers, d3-GHR homodimers and full-length GHR–d3-GHR heterodimers. Because of its prominent function in growth hormone signaling and the frequency of its polymorphic variation, we considered GHR to be a good candidate gene for involvement in the pharmacogenomics of growth hormone therapy.
We studied the effect of the exon 3 polymorphism in GHR on response to growth hormone therapy in a large sample of short children. We selected children born small for gestational age (SGA) and children with idiopathic short stature (ISS), who have normal birth size but grow at a decreased rate1,2,3. During the study, the children were in the phase of linear growth before the onset of puberty; this allowed us to avoid the extensive variations in growth related to individual patterns of sexual maturation. We recruited two independent cohorts of ISS and SGA children with short stature (Table 1) and genotyped them for the GHR polymorphism. The frequency of the d3-GHR variant was comparable in the short children and in 283 control adults of normal height, suggesting that this polymorphism is not primarily related to the genesis of short stature in individuals with ISS and SGA. GHR genotype had no significant effect on growth rates or hormonal parameters before therapy (Table 1). We concluded from this data that potential variations in growth hormone sensitivity due to GHR differences in short children can be compensated by endogenous pituitary growth hormone secretion, which masks the effects of the GHR polymorphism on basal growth rate. Unfortunately, growth hormone stimulation tests used in clinical endocrinology are unreliable reflections of endogenous growth hormone secretion and did not allow us to evaluate this hypothesis properly.
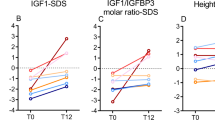
Our primary question was whether response to growth hormone differed across GHR genotypes. We studied children with SGA and ISS during the first two years of growth hormone administration. The distribution of individual growth responses, indicating various degrees of sensitivity to growth hormone, closely fit that of the normal distribution, as expected for a multifactorial trait (Fig. 1). But the distribution of genotypes across the range of growth acceleration values indicated that the growth responses to growth hormone were greater in children bearing at least one allele encoding the d3-GHR isoform. The unadjusted mean growth responses stratified by GHR genotype are shown in Table 2. In the first sample of 76 short children, GHR genotype had a significant effect on response to growth hormone during the two years of growth hormone administration (P < 10−5 and P < 0.001 for the first and second years, respectively). Response to growth hormone was greater in children carrying at least one allele encoding the d3-GHR isoform. To further characterize the effect of the allele encoding the d3-GHR isoform on response to growth hormone, we built a general linear regression model taking into account age, sex and growth hormone dose (Table 3). We found a strong relationship between GHR genotype and growth acceleration (P < 10−6 and P < 0.005 for the first and second years of growth hormone administration, respectively; Table 2). The regression model also showed the expected relationship between growth hormone dose and growth response, which was attenuated during the second year of growth hormone administration because many subjects were switched from a fixed 'per protocol' growth hormone dose (in the first year) to a personalized dose regimen (in the second year).
The distribution of values was normal and had a large variance. The two cohorts of short children treated with growth hormone were pooled for analysis. Increments in growth rates were greater in children carrying alleles encoding the d3-GHR isoform. fl, allele encoding the full-length GHR isoform; d3, allele encoding the d3-GHR isoform.
We replicated these findings in a second, independent sample of 96 short children (Table 1). Response to growth hormone in this sample differed across GHR genotypes (Table 1), comparable to that in the first sample (Table 2). As expected, we observed no differences in the effects of GHR genotype between children with SGA and children with ISS.
To verify that the observed effect could be reliably attributed to GHR, we examined nine polymorphisms of high heterozygosity in genes unlinked to GHR. None showed evidence of association with treatment response or GHR genotypes in our cohorts. Because we carefully matched the children with SGA and ISS with respect to ethnic background, the observed association is probably not due to the effects of population stratification.
We then investigated in vitro whether the deletion of exon 3 affects the functional properties of GHR. We transiently cotransfected 293 HEK fibroblasts with vectors expressing full-length GHR, d3-GHR or both using the LHRE-luciferase reporter plasmid, which is activated by full-length GHR2,11. When cells were exposed to various growth hormone concentrations, d3-GHR induced a higher transcriptional activity of the reporter construct than full-length GHR (Fig. 2). The results were linear and consistent at all levels of growth hormone stimulation between 0 and 50 ng ml−1, a range of low growth hormone concentrations supporting the in vivo relevance of our findings. 293 HEK fibroblasts expressing d3-GHR had a greater response to growth hormone stimulation in all our experiments (Fig. 2). To our knowledge, this is the first report comparing the bioactivity of full-length GHR and d3-GHR in vitro. The molecular mechanisms underlying the higher bioactivity of d3-GHR are not yet elucidated, but they do not seem to involve more binding7,8,9 or less internalization8. In vivo, full-length GHR and d3-GHR isoforms might be discriminated at the level of expression in lymphocytes12. This is not the case in our assay, as both receptors are under the control of the constitutively active CMV promoter.
HEK 293 cells transiently expressing full-length GHR, d3-GHR or both were stimulated by increasing concentrations of growth hormone ([GH]) for 8 h. Relative induction of LHRE-luciferase reporter gene is expressed relative to unstimulated cells (value of 1, horizontal line). The number of experiments done for each condition is indicated in parentheses. *P < 0.005, **P < 0.0005 and ***P < 0.0001 for comparison of cells transfected with two alleles encoding the full-length GHR isoform versus cells transfected with at least one allele encoding the d3-GHR isoform. We pooled cells carrying one and two alleles encoding the d3-GHR isoform because their individual effects were each consistently higher than those of cells carrying two alleles encoding the full-length GHR isoform (P = 0.006–0.0005 and P = 0.004–0.0001, respectively). fl, allele encoding the full-length GHR isoform; d3, allele encoding the d3-GHR isoform.
Recent studies showed that growth hormone receptors exist at the cell membrane as inactive, preformed dimers13 whose activation is triggered by conformational changes induced by growth hormone binding, possibly involving the rotation of receptor chains (M. J. Waters et al., unpublished data). The first 32 residues (including residues 7–28 encoded by exon 3) could not be modeled in any of the available crystal structures involving GHR6,14, suggesting that the N-terminal loop has a high degree of flexibility. It is not known how the deletion of the region encoded by exon 3 could increase receptor activity, but removal of this flexible loop may propagate subtle conformational changes along the extracellular domain that facilitate hormone-triggered activation of the receptor.
In 2001 and in 2003, the US Food and Drug Administration and the European regulatory agency, respectively, allowed the treatment of short children born SGA with recombinant growth hormone. In July 2003, growth hormone was also approved by the US Food and Drug Administration for the treatment of growth failure in children with ISS. Children whose height is 2.25–3 s.d. below the mean for their ages are eligible for growth hormone prescription; this includes hundreds of thousands of children in the US and Europe. For those short children, pharmacogenomic testing could be used in the future to avoid the use of ineffective regimens of growth hormone, to improve the personalized design of therapy and to decrease its cost:benefit ratio. Our study on GHR polymorphism is a preliminary step in this direction. If it holds true, our observation will have more useful consequences for the therapy of short children than for direct diagnostic purposes, because although it is involved in the multifactorial network of growth genetics, the exon 3 variation in GHR is not expected to be associated with physiologic variations in human growth or to be a primary cause of short stature in humans. This is because individual differences in growth hormone sensitivity due to GHR structure and function are probably easily compensated for by pituitary growth hormone secretion in a secretion-sensitivity balance that allows optimal growth. When this compensation is impossible, however, as in growth hormone–deficient children, or limited by its own genetic variation, homozygosity with respect to the allele encoding full-length GHR could aggravate the growth deficit associated with low growth hormone secretion.
The main finding of our study is that, in response to a given dose of exogenous growth hormone, and regardless of the cause of shortness, individual differences in GHR genotype have medically relevant effects on growth rate. Prospective dose-response studies in all categories of children treated with growth hormone will determine the optimal growth hormone dosage for their GHR genotype. This may contribute to the progressive switch in growth hormone prescription from a 'fixed dosage' mode of therapy, which prevails now, to a more personalized adjustment of dose.
Methods
Individuals studied.
We recruited two cohorts of 76 and 96 children of European descent with stature more than 2 s.d. below the mean who were selected according to criteria for SGA and ISS1,3. We determined European ancestry on the basis of the patronymic and geographic origin of each child's four grandparents. SGA describes a neonate whose crown-heel length at birth is at least 2 s.d. below the mean for the infant's gestational age. All children were fully evaluated with two growth hormone stimulation tests, measurement of IGF1, thyroid hormones and TSH, karyotype, skeleton X-ray and careful clinical examination by pediatric endocrinologists (P.B., C.T., M.T.). None of these short children had deficiencies of growth hormone or other hormones, any chromosomal abnormality, dysmorphic syndrome, skeletal disease or any other medical condition. Selected children were treated with different doses of growth hormone (Table 1) and were measured every 2–3 months thereafter at our centers. Bone age was comparable in all genotypic subgroups at initiation of therapy. All measurements of height were done in duplicate by trained nurses using a Harpenden stadiometer. Written consent was obtained according to the French Bioethics law and our Institutional Review Board of Paris 5. We also obtained samples from 283 controls of normal height.
We did not carry out this study using children deficient in growth hormone because the causes for this deficiency are highly heterogeneous (mutations of pituitary transcription factors, irradiation for cancer, hypothalamic tumors, etc.) and are often associated with other hormone deficits or medical problems. A separate study of growth hormone pharmacogenomics in non-tumor-related growth hormone deficiency is underway.
Genotyping.
To determine genotype at GHR exon 3, we designed a simple multiplex PCR assay4, using primers G1, G2 and G3 (GenBank accession number AF155912), as follows: initial step of denaturation of 5 min at 94 °C, followed by 35 cycles consisting of 30 s at 94 °C, 30 s at 60 °C and 1 min and 30 s at 72 °C, followed by an extension period at 72 °C for 7 min. We analyzed the amplification products by electrophoresis on a 1% agarose gel stained with ethidium bromide. Allele frequencies were in Hardy-Weinberg equilibrium and were comparable in our children with SGA or IDSS and in 283 unaffected subjects of normal adult height.
Genomic control.
To safeguard against population stratification in children carrying two alleles encoding full-length GHR (poor responders) versus those carrying at least one allele encoding d3-GHR (good responders), we included only individuals of European origin as determined based on birthplace and patronymics of the four grandparents.
In addition, we examined the selected children for nine markers in genes that are unlinked to GHR, some of which are known to have considerable variation between human populations: INS (11p15.5; ref. 15), ADRB2 (5q31–q32; ref. 16), LHCGR (2p21), PIK3CB (3q22.3), KCNJ11 (11p15.1), ADRA2A (10q24–q26), ADRB3 (8p12–p11.2), LEP (7q31.3) and UCP2 (11q13; data not shown). We examined these markers for association with the growth rate in response to growth hormone; such associations would be indicative of trait stratification. The ANOVA results were not significant in all instances (nominal P > 0.05). We also examined genotype differences at these loci between the good responders and the poor responders to test for evidence of population stratification associated with the GHR genotype. Again, the nominal P values were not significant. Therefore, from the available genomic control data, we conclude that there is no evidence for population stratification in the cohort.
Calculations and statistical methods.
The growth rate response to growth hormone administration was expressed as Δgr, the difference between growth rate during growth hormone treatment (in cm y−1) and previous growth rate (evaluated during the whole year before onset of growth hormone administration). Height was measured in standing position with a Harpenden stadiometer that was always the same for all measurements in the studied children. We chose this quantitative criteria because spontaneous growth in children with SGA and ISS is linear during this period of childhood1,2. We found that Δgr was more precise than a Z score because precise growth curves were available for all the children we studied. We did not study the response of circulating IGF1 to growth hormone administration in bona fide conditions, because children were not sampled in a codified nutritional status. The intra-individual values for IGF1 were too variable during growth hormone therapy to be considered reliable indices of growth hormone sensitivity.
Wilcoxon rank tests and unpaired Student's t-tests were both used to compare the results of transfections with full-length GHR or d3-GHR (Fig. 2). We fitted a multivariate linear model to the data to estimate the proper effect of the tested GHR polymorphism on response to growth hormone, adjusted for the effect of the other covariates contributing to the growth under treatment, such as age at diagnosis, sex and the received dose of growth hormone. This approach is suitable for estimating the association between the variable of interest, here the genetic polymorphism, and the trait in the presence of correlation between the covariates. We carried out tests of independence of each covariate one at a time, keeping the others in the model. Statistics and estimations of effect given in the tables are thus adjusted for the others and are not subject to marginal association. We checked the normality of the residuals, and the residuals versus the fitted values did not show any trend, indicating that there was no noticeable deviations from the assumption of the linear model. All linear model and statistics in Table 1 were computed using R v1.6.0 (ref. 17).
GHR and Stat5 reporter constructs.
We amplified DNA fragments of 1,921 bp and 1,855 bp from one of the two human GHR cDNAs, encoding full-length GHR and d3-GHR, respectively (primer sequences are available on request). We separated these fragments by electrophoresis on a 1% agarose gel, cut them and purified them using the Qiaquick Gel Extraction kit (Qiagen). We then inserted each fragment directly into pcDNA3.1/V5-His-TOPO vector plasmid (Invitrogen). All constructs were checked by DNA sequencing, which confirmed that retention or exclusion of exon 3 was the only difference between the two amplified fragments. The growth hormone–responsive reporter gene encodes the firefly luciferase gene under the control of an artificial promoter containing six tandem repeats of STAT5 response element, referred to as lactogenic hormone response element (LHRE), and the minimal thymidine kinase promoter11,18. We purchased the plasmid encoding Renilla luciferase (pRL-TK plasmid) from Promega.
Transient transfections and luciferase assays.
We plated 5 × 104 293 cells on 96-well plates of the appropriate growth medium supplemented with 10% fetal calf serum and transfected them with vector plasmid (20 ng per well) encoding full-length GHR or d3-GHR alone or together (1:1 ratio) using Fugene (Roche). We prepared five different DNA preparations for each plasmid. We cotransfected cells in each well with LHRE-luciferase and Renilla luciferase plasmids (4 ng per well) to normalize the growth hormone–induced firefly luciferase activity. Sixteen hours after transfection, we stimulated cells for 8 h with different concentrations of recombinant human growth hormone (Saizen, Serono). We determined luciferase activities using the dual-luciferase reporter assay system as indicated by the manufacturer (Promega) using a Centro LB 960 luminometer (Berthold). Normalized luciferase (firefly versus Renilla) data are expressed as relative induction (stimulated versus nonstimulated cells).
Accession codes
References
Lesage, C., Walker, J., Landier, F., Chatelain, P. & Bougnères, P.F. Near normalization of adolescent height with growth hormone therapy in very short children without growth hormone deficiency. J. Pediatr. 119, 29–34 (1991).
Hintz, R.L., Attie, K.M., Baptista, J. & Roche, A. Effect of growth hormone treatment on adult height of children with idiopathic short stature. Genentech Collaborative Group. N. Engl. J. Med. 340, 502–507 (1999).
Bryant, J., Cave, C. & Milne, R. Recombinant growth hormone for idiopathic short stature in children and adolescents. Cochrane Database Syst. Rev. CD004440 (2003).
Pantel, J. et al. Species-specific alternative splice mimicry at the growth hormone receptor locus revealed by the lineage of retroelements during primate evolution. J. Biol. Chem. 275, 18664–18669 (2000).
Kelly, P.A. et al. The growth hormone/prolactin receptor family. Recent Prog. Horm. Res. 48, 123–164 (1993).
de Vos, A.M., Ultsch, M. & Kossiakoff, A.A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255, 306–312 (1992).
Sobrier, M.L., Duquesnoy, P., Duriez, B., Amselem, S. & Goossens, M. Expression and binding properties of two isoforms of the human growth hormone receptor. FEBS Lett. 319, 16–20 (1993).
Urbanek, M., Russell, J.E., Cooke, N.E. & Liebhaber, S.A. Functional characterization of the alternatively spliced, placental human growth hormone receptor. J. Biol. Chem. 268, 19025–19032 (1993).
Bass, S.H., Mulkerrin, M.G. & Wells, J.A. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc. Natl. Acad. Sci. USA 88, 4498–4502 (1991).
Pantel, J. et al. Heterozygous nonsense mutation in exon 3 of the growth hormone receptor (GHR) in severe GH insensitivity (Laron syndrome) and the issue of the origin and function of the GHRd3 isoform. J. Clin. Endocrinol. Metab. 88, 1705–1710 (2003).
Sotiropoulos, A. et al. Differential activation of Stat3 and Stat5 by distinct regions of the growth hormone receptor. Mol. Endocrinol. 10, 998–1009 (1996).
Ochoa, R. et al. Expression of growth hormone receptor isoform exon-3-excluding and exon-3-retaining messenger RNAs in peripheral lymphocytes from normal and acromegalic subjects. Horm. Res. 60, 68–72 (2003).
Frank, S.J. Receptor dimerization in GH and erythropoietin action—it takes two to tango, but how? Endocrinology 143, 2–10 (2002).
Sundstrom, M. et al. Crystal structure of an antagonist mutant of human growth hormone, G120R, in complex with its receptor at 2.9 A resolution. J. Biol. Chem. 271, 32197–32203 (1996).
Stead, J.D. & Jeffreys, A.J. Allele diversity and germline mutation at the insulin minisatellite. Hum. Mol. Genet. 9, 713–723 (2000)
Drysdale, C.M., et al. Complex promoter and coding region beta-2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. 97, 10483–10488 (2000).
Ithaka, R. & Gentelman, R.R. A language for data analysis and graphics. J. Comp. Graph. Stats. 5, 299–314 (1996).
Goffin, V. et al. Antagonistic properties of human prolactin analogs that show paradoxical agonistic activity in the Nb2 bioassay. J. Biol. Chem. 271, 16573–16579 (1996).
Acknowledgements
We thank M. Lathrop for carrying out the statistical tests for genomic control, F. Suarez and C. Molinas for their help, C. LeStunff and Shalimar for daily support in the laboratory and P. Chatelain and M. L. Sobrier for discussions. We thank Pfizer for supporting our Growth Genomics Project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Dos Santos, C., Essioux, L., Teinturier, C. et al. A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nat Genet 36, 720–724 (2004). https://doi.org/10.1038/ng1379
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1379
This article is cited by
-
The in vitro functional analysis of single-nucleotide polymorphisms associated with growth hormone (GH) response in children with GH deficiency
The Pharmacogenomics Journal (2019)
-
Expression of the growth hormone receptor isoforms and its correlation with the metabolic profile in morbidly obese subjects
Endocrine (2019)
-
The P2 promoter of the IGF1 gene is a major epigenetic locus for GH responsiveness
The Pharmacogenomics Journal (2016)
-
Growth hormone receptor exon 3 isoforms may have no importance in the clinical setting of multiethnic Brazilian acromegaly patients
Pituitary (2016)
-
Clinical features and natural course of acromegaly in patients with discordance in the nadir GH level on the oral glucose test and the IGF-1 value at 3 months after adenomectomy
Neurosurgical Review (2016)