Abstract
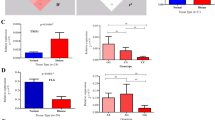
Psoriasis is a multifactorial skin disease characterized by epidermal hyperproliferation and chronic inflammation, the most common form of which is psoriasis vulgaris (PsV). We present a genome-wide association analysis of 2,339,118 SNPs in 472 PsV cases and 1,146 controls from Germany, with follow-up of the 147 most significant SNPs in 2,746 PsV cases and 4,140 controls from three independent replication panels. We identified an association at TRAF3IP2 on 6q21 and genotyped two SNPs at this locus in two additional replication panels (the combined discovery and replication panels consisted of 6,487 cases and 8,037 controls; combined P = 2.36 × 10−10 for rs13210247 and combined P = 1.24 × 10−16 for rs33980500). About 15% of psoriasis cases develop psoriatic arthritis (PsA). A stratified analysis of our datasets including only PsA cases (1,922 cases compared to 8,037 controls, P = 4.57 × 10−12 for rs33980500) suggested that TRAF3IP2 represents a shared susceptibility for PsV and PsA. TRAF3IP2 encodes a protein involved in IL-17 signaling and which interacts with members of the Rel/NF-κB transcription factor family.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
References
Griffiths, C.E. & Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 370, 263–271 (2007).
Bowcock, A.M. & Barker, J.N. Genetics of psoriasis: the potential impact on new therapies. J. Am. Acad. Dermatol. 49, S51–S56 (2003).
Gottlieb, A.B. Psoriasis: emerging therapeutic strategies. Nat. Rev. Drug Discov. 4, 19–34 (2005).
Oudot, T. et al. An association study of 22 candidate genes in psoriasis families reveals shared genetic factors with other autoimmune and skin disorders. J. Invest. Dermatol. 129, 2637–2645 (2009).
Bhalerao, J. & Bowcock, A.M. The genetics of psoriasis: a complex disorder of the skin and immune system. Hum. Mol. Genet. 7, 1537–1545 (1998).
Elder, J.T. et al. The genetics of psoriasis. Arch. Dermatol. 130, 216–224 (1994).
Tsunemi, Y. et al. Interleukin-12 p40 gene (IL12B) 3-untranslated region polymorphism is associated with susceptibility to atopic dermatitis and psoriasis vulgaris. J. Dermatol. Sci. 30, 161–166 (2002).
Cargill, M. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
Capon, F. et al. Sequence variants in the genes for the interleukin-23 receptor (IL23R) and its ligand (IL12B) confer protection against psoriasis. Hum. Genet. 122, 201–206 (2007).
Nair, R.P. et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J. Invest. Dermatol. 128, 1653–1661 (2008).
Liu, Y. et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 4, e1000041 (2008).
Chang, M. et al. Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes Immun. 9, 176–181 (2008).
Nair, R.P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41, 199–204 (2009).
de Cid, R. et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 41, 211–215 (2009).
Zhang, X.J. et al. Polymorphisms in interleukin-15 gene on chromosome 4q31.2 are associated with psoriasis vulgaris in Chinese population. J. Invest. Dermatol. 127, 2544–2551 (2007).
Sherry, S.T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Ye, H. et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418, 443–447 (2002).
Darnay, B.G., Ni, J., Moore, P.A. & Aggarwal, B.B. Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-kappaB-inducing kinase. Identification of a novel TRAF6 interaction motif. J. Biol. Chem. 274, 7724–7731 (1999).
Gelfand, J.M. et al. Epidemiology of psoriatic arthritis in the population of the United States. J. Am. Acad. Dermatol. 53, 573 (2005).
Zhou, X. et al. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol. Genomics 13, 69–78 (2003).
Qian, Y. et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 8, 247–256 (2007).
Qian, Y. et al. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Immunity 21, 575–587 (2004).
Lowes, M.A. et al. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J. Invest. Dermatol. 128, 1207–1211 (2008).
Kryczek, I. et al. Induction of IL-17+ T cell trafficking and development by IFN-γ: mechanism and pathological relevance in psoriasis. J. Immunol. 181, 4733–4741 (2008).
Zaba, L.C. et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J. Allergy Clin. Immunol. 124, 1022–10 e1–395 (2009).
Hunter, C.A. Act1-ivating IL-17 inflammation. Nat. Immunol. 8, 232–234 (2007).
Moll, J.M. & Wright, V. Psoriatic arthritis. Semin. Arthritis Rheum. 3, 55–78 (1973).
Taylor, W. et al. Classification criteria for psoriatic arthritis: development of new criteria from a large international study. Arthritis Rheum. 54, 2665–2673 (2006).
Krawczak, M. et al. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 9, 55–61 (2006).
Wichmann, H.E., Gieger, C. & Illig, T. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen 67 Suppl 1, S26–S30 (2005).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
The International HapMap Consortium. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Dupont, W.D. & Plummer, W.D. PS power and sample size program available for free on the Internet. Control. Clin. Trials 18 (1997).
Willer, C.J., Li, Y. & Abeçasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Stoll, S.W., Johnson, J.L., Li, Y., Rittie, L. & Elder, J.T. Amphiregulin carboxy-terminal domain is required for autocrine keratinocyte growth. J. Invest. Dermatol. 130, 2031–2040 (2010).
Stoll, S.W. et al. Metalloproteinase-mediated, context-dependent function of amphiregulin and HB-EGF in human keratinocytes and skin. J. Invest. Dermatol. 130, 295–304 (2010).
Acknowledgements
We thank all individuals with psoriasis who participated in this study, their families and their physicians for their cooperation. We acknowledge the cooperation of Genizon Biosciences. We wish to thank T. Wesse, T. Henke, C. Fürstenau, S. Ehlers, M. Davids and R. Vogler for expert technical help. We thank J.R. Hov for helpful discussions. This study was supported by the German Ministry of Education and Research (BMBF) through the National Genome Research Network (NGFN), the PopGen biobank and the KORA (Cooperative Research in the Region of Augsburg) research platform. KORA was initiated and financed by the Helmholtz Zentrum München-National Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria and the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. The project received infrastructure support through the Deutsche Forschungsgemeinschaft (DFG) clusters of excellence 'Multimodal Computing and Interaction' and 'Inflammation at Interfaces'. This research was also supported by grants R01-AR42742, R01-AR050511 and R01-AR054966 from the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
E.E. performed SNP selection, genotyping, data analysis and prepared the figures and tables. A.F. helped with data analysis. D.E. performed imputation, data analysis and generated the regional plots. P.E.S., J.G., J.D. and Y.L. performed the expression and expression quantitative trait loci analyses. S.W.S. and S.L. performed small hairpin RNA (shRNA) experiments. G.R.A. helped with statistical analyses and interpretation of the results. M.A. and G.M. performed in silico protein analyses. M.W., U.M., S.W., B.E. and M.K. coordinated the recruitment and collected phenotype data of panels A and B. C.G. and H.E.W. provided the KORA control samples. J.T.E., J.J.V., R.P.N., T.T., S.D., J.V.R., M.B., H.F., C.R., P.R. and D.D.G. provided the replication samples C through F and the respective genotypes and phenotypes. T.H.K., R.P.N. and D.K. helped with genotyping. E.E. and A.F. drafted the manuscript. D.E., M.W., J.T.E., T.H.K. and S.S. edited the manuscript. A.F. planned and supervised the study. All authors approved the final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3. (PDF 2989 kb)
Rights and permissions
About this article
Cite this article
Ellinghaus, E., Ellinghaus, D., Stuart, P. et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet 42, 991–995 (2010). https://doi.org/10.1038/ng.689
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.689
This article is cited by
-
The use of leukocytes’ secretome to individually target biological therapy in autoimmune arthritis: a case report
Clinical and Translational Medicine (2019)
-
The Act1 D10N missense variant impairs CD40 signaling in human B-cells
Genes & Immunity (2019)
-
New polymorphisms associated with response to anti-TNF drugs in patients with moderate-to-severe plaque psoriasis
The Pharmacogenomics Journal (2018)
-
Axial disease in psoriatic arthritis and ankylosing spondylitis: a critical comparison
Nature Reviews Rheumatology (2018)
-
Anti-inflammatory Effects of Atorvastatin by Suppressing TRAF3IP2 and IL-17RA in Human Glioblastoma Spheroids Cultured in a Three-dimensional Model: Possible Relevance to Glioblastoma Treatment
Molecular Neurobiology (2018)