Abstract
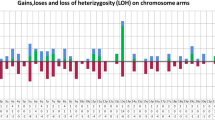
Recent advances in genome-wide single-nucleotide polymorphism (SNP) analyses have revealed previously unrecognized microdeletions and uniparental disomy (UPD) in a broad spectrum of human cancers. As acute myeloid leukemia (AML) represents a genetically heterogeneous disease, this technology might prove helpful, especially for cytogenetically normal AML (CN-AML) cases. Thus, we performed high-resolution SNP analyses in 157 adult cases of CN-AML. Regions of acquired UPDs were identified in 12% of cases and in the most frequently affected chromosomes, 6p, 11p and 13q. Notably, acquired UPD was invariably associated with mutations in nucleophosmin 1 (NPM1) or CCAAT/enhancer binding protein-α (CEBPA) that impair hematopoietic differentiation (P=0.008), suggesting that UPDs may preferentially target genes that are essential for proliferation and survival of hematopoietic progenitors. Acquired copy number alterations (CNAs) were detected in 49% of cases with losses found in two or more cases affecting, for example, chromosome bands 3p13–p14.1 and 12p13. Furthermore, we identified two cases with a cryptic t(6;11) as well as several non-recurrent aberrations pointing to leukemia-relevant regions. With regard to clinical outcome, there seemed to be an association between UPD 11p and UPD 13q cases with overall survival. These data show the potential of high-resolution SNP analysis for identifying genomic regions of potential pathogenic and clinical relevance in AML.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Estey E, Dohner H . Acute myeloid leukaemia. Lancet 2006; 368: 1894–1907.
Grimwade D . The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol 2001; 14: 497–529.
Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD . Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood 2007; 109: 431–448.
Dohner K, Dohner H . Molecular characterization of acute myeloid leukemia. Haematologica 2008; 93: 976–982.
Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008; 358: 1909–1918.
Carter NP . Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet 2007; 39 (7 Suppl): S16–S21.
Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, Zhang J et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res 2006; 16: 1575–1584.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–114.
Frohling S, Dohner H . Chromosomal abnormalities in cancer. N Engl J Med 2008; 359: 722–734.
Raghavan M, Lillington DM, Skoulakis S, Debernardi S, Chaplin T, Foot NJ et al. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res 2005; 65: 375–378.
Gupta M, Raghavan M, Gale RE, Chelala C, Allen C, Molloy G et al. Novel regions of acquired uniparental disomy discovered in acute myeloid leukemia. Genes Chromosomes Cancer 2008; 47: 729–739.
Fitzgibbon J, Smith LL, Raghavan M, Smith ML, Debernardi S, Skoulakis S et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res 2005; 65: 9152–9154.
Gondek LP, Tiu R, O′Keefe CL, Sekeres MA, Theil KS, Maciejewski JP . Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood 2008; 111: 1534–1542.
Mohamedali A, Gaken J, Twine NA, Ingram W, Westwood N, Lea NC et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood 2007; 110: 3365–3373.
Gaidzik VI, Schlenk RF, Moschny S, Becker A, Bullinger L, Corbacioglu A et al. Prognostic impact of WT1 mutations in cytogenetically normal acute myeloid leukemia (AML): a study of the German-Austrian AML Study Group (AMLSG). Blood 2009; 113: 4505–4511.
Pounds S, Cheng C, Mullighan C, Raimondi SC, Shurtleff S, Downing JR . Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics 2009; 25: 315–321.
Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res 2005; 65: 6071–6079.
Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of Affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet 2007; 81: 114–126.
Frohling S, Kayser S, Mayer C, Miller S, Wieland C, Skelin S et al. Diagnostic value of fluorescence in situ hybridization for the detection of genomic aberrations in older patients with acute myeloid leukemia. Haematologica 2005; 90: 194–199.
R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing: Vienna 2007.
Suela J, Alvarez S, Cifuentes F, Largo C, Ferreira BI, Blesa D et al. DNA profiling analysis of 100 consecutive de novo acute myeloid leukemia cases reveals patterns of genomic instability that affect all cytogenetic risk groups. Leukemia 2007; 21: 1224–1231.
Maciejewski JP, Mufti GJ . Whole genome scanning as a cytogenetic tool in hematologic malignancies. Blood 2008; 112: 965–974.
Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA 2009; 106: 12950–12955.
Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y et al. Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD et al. Global variation in copy number in the human genome. Nature 2006; 444: 444–454.
Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P et al. Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525–528.
Rucker FG, Bullinger L, Schwaenen C, Lipka DB, Wessendorf S, Frohling S et al. Disclosure of candidate genes in acute myeloid leukemia with complex karyotypes using microarray-based molecular characterization. J Clin Oncol 2006; 24: 3887–3894.
Novak RL, Phillips AC . Adenoviral-mediated Rybp expression promotes tumor cell-specific apoptosis. Cancer Gene Ther 2008; 15: 713–722.
Streubel B, Vinatzer U, Lamprecht A, Raderer M, Chott A . T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 2005; 19: 652–658.
Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 2002; 100: 4325–4336.
Tyybakinoja A, Elonen E, Piippo K, Porkka K, Knuutila S . Oligonucleotide array-CGH reveals cryptic gene copy number alterations in karyotypically normal acute myeloid leukemia. Leukemia 2007; 21: 571–574.
Akagi T, Ogawa S, Dugas M, Kawamata N, Yamamoto G, Nannya Y et al. Frequent genomic abnormalities in acute myeloid leukemia/myelodysplastic syndrome with normal karyotype. Haematologica 2009; 94: 213–223.
Barjesteh van Waalwijk van Doorn-Khosrovani S, Spensberger D, de Knegt Y, Tang M, Lowenberg B, Delwel R . Somatic heterozygous mutations in ETV6 (TEL) and frequent absence of ETV6 protein in acute myeloid leukemia. Oncogene 2005; 24: 4129–4137.
Bignone PA, Lee KY, Liu Y, Emilion G, Finch J, Soosay AE et al. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer. Oncogene 2007; 26: 683–700.
Asou N, Yanagida M, Huang L, Yamamoto M, Shigesada K, Mitsuya H et al. Concurrent transcriptional deregulation of AML1/RUNX1 and GATA factors by the AML1-TRPS1 chimeric gene in t(8;21)(q24;q22) acute myeloid leukemia. Blood 2007; 109: 4023–4027.
Lapointe J, Malhotra S, Higgins JP, Bair E, Thompson M, Salari K et al. hCAP-D3 expression marks a prostate cancer subtype with favorable clinical behavior and androgen signaling signature. Am J Surg Pathol 2008; 32: 205–209.
Uht RM, Amos S, Martin PM, Riggan AE, Hussaini IM . The protein kinase C-eta isoform induces proliferation in glioblastoma cell lines through an ERK/Elk-1 pathway. Oncogene 2007; 26: 2885–2893.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009; 360: 2289–2301.
Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 2009; 41: 838–842.
Corbacioglu A, Fröhling S, Paschka P, Marcucci G, Anhalt A, Urlbauer K et al. Acute myeloid leukemia (AML) with 9q aberrations occurring within a non-complex karyotype is highly associated with CEBPA and NPM1 mutations - a joint analysis of the German-Austrian AML Study Group (AMLSG) and Cancer and Leukemia Group B (CALGB). Blood (ASH Annu Meet Abstr) 2007; 110: 762.
Frohling S, Schlenk RF, Krauter J, Thiede C, Ehninger G, Haase D et al. Acute myeloid leukemia with deletion 9q within a noncomplex karyotype is associated with CEBPA loss-of-function mutations. Genes Chromosomes Cancer 2005; 42: 427–432.
Peniket A, Wainscoat J, Side L, Daly S, Kusec R, Buck G et al. Del (9q) AML: clinical and cytological characteristics and prognostic implications. Br J Haematol 2005; 129: 210–220.
Sweetser DA, Peniket AJ, Haaland C, Blomberg AA, Zhang Y, Zaidi ST et al. Delineation of the minimal commonly deleted segment and identification of candidate tumor-suppressor genes in del(9q) acute myeloid leukemia. Genes Chromosomes Cancer 2005; 44: 279–291.
Moumen A, Masterson P, O′Connor MJ, Jackson SP . hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 2005; 123: 1065–1078.
Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS . Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature 2004; 430: 1034–1039.
Aplan PD . Chromosomal translocations involving the MLL gene: molecular mechanisms. DNA Repair (Amst) 2006; 5: 1265–1272.
Marlton P, Claxton DF, Liu P, Estey EH, Beran M, LeBeau M et al. Molecular characterization of 16p deletions associated with inversion 16 defines the critical fusion for leukemogenesis. Blood 1995; 85: 772–779.
Radtke I, Mullighan CG, Ishii M, Su X, Cheng J, Ma J et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci USA 2009; 106: 12944–12949.
Falini B, Nicoletti I, Martelli MF, Mecucci C . Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood 2007; 109: 874–885.
Acknowledgements
For excellent technical assistance we thank Karina Eiwen, Sabrina Heinrich, Marianne Habdank, Karin Lanz and Simone Miller. Thanks also to Dr Jing Ma und Dr Xiaoping Su for their help with data analysis, and we thank all the members of the German-Austrian AMLSG for their participation in the clinical trials and for providing leukemia samples. This study was supported in part by the Deutsche Forschungsgemeinschaft (SCHO 1296/1-1), the Else Kröner-Fresenius-Stiftung (P32/2004) and the American Lebanese and Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital. Lars Bullinger designed and performed the research, analyzed/interpreted the data and wrote the paper; Jan Krönke performed the research and analyzed/interpreted the data; Christian Schön performed the research and analyzed/interpreted the data; Ina Radtke analyzed/interpreted the data; Katja Urlbauer performed the research and analyzed the data; Ursula Botzenhardt, Verena Gaidzik, Ana Carió and Christine Senger performed the research; Richard F Schlenk analyzed/interpreted the data; James R Downing contributed the analytical tools and analyzed/interpreted the data; Karlheinz Holzmann designed and performed the research and analyzed/interpreted the data; Konstanze Döhner designed the research, analyzed/interpreted the data and wrote the paper; Hartmut Döhner designed the research, contributed vital reagents, analyzed/interpreted the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Rights and permissions
About this article
Cite this article
Bullinger, L., Krönke, J., Schön, C. et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 24, 438–449 (2010). https://doi.org/10.1038/leu.2009.263
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2009.263
Keywords
This article is cited by
-
Novel prognostic genes and subclasses of acute myeloid leukemia revealed by survival analysis of gene expression data
BMC Medical Genomics (2021)
-
HLA-haplotype loss after TCRαβ/CD19-depleted haploidentical HSCT
Bone Marrow Transplantation (2021)
-
Identifying a novel 5-gene signature predicting clinical outcomes in acute myeloid leukemia
Clinical and Translational Oncology (2021)
-
The transcription factor Rfx7 limits metabolism of NK cells and promotes their maintenance and immunity
Nature Immunology (2018)
-
Clinical implications of genome-wide DNA methylation studies in acute myeloid leukemia
Journal of Hematology & Oncology (2017)