Abstract
The relationship of TNF-α promoter polymorphisms and ankylosing spondylitis (AS) has been reported with conflicting results. We perform this meta-analysis to collect all the relevant studies up to date to further clarify the association of TNF-α promoter polymorphisms with AS. A review was conducted of studies reporting on the association between TNF-α promoter polymorphisms and AS susceptibility in Medline, Pubmed, Embase, and Web of Science. The numbers of individuals with various genotypes and alleles in both the case and control groups were extracted from relevant studies. Odds ratios (ORs) with 95% confidence interval (CI) were used to estimate the association. Fourteen eligible studies, contributing data on 3,880 subjects (1,766 patients; 2,114 controls), were included in this meta-analysis. The ORs of various comparisons indicated that there was no association between TNF-α 238, 308 polymorphisms, and AS susceptibility in the overall population. For HLA-B27+ population, although the frequency of 308 A allele decreased in AS patients (OR = 0.721; 95%CI = 0.522–0.995), the result was no longer statistically significant after excluding the Hardy–Weinberg equilibrium violation studies (OR = 1.150; 95%CI = 0.568–2.310). No relationship was found between TNF-α promoter 238 polymorphisms and AS in HLA-b27+ population. No association was found between TNF-α promoter 238/308 polymorphisms and ankylosing spondylitis susceptibility in both the overall and HLA-B27+ population.
Similar content being viewed by others
Introduction
Ankylosing spondylitis (AS) is a chronic inflammatory spondyloarthropathy which is characterized by enthesitis, axial, and peripheral arthritis. As a class I antigen of the major histocompatibility complex (MHC) system, human leukocyte antigen (HLA)-B27 has been proven to be strongly associated with the susceptibility of AS [1, 2]. However, increasing evidence indicated that other genetic factors were also involved in AS susceptibility except HLA-B27 [3–5].
Tumor necrosis factor alpha (TNF-α), as a macrophage-derived pro-inflammatory cytokine, has been recently implicated in the pathogenesis and development of AS [6]. The TNF-α gene is 12 kb in length and located in class III region of MHC, adjacent to the HLA-B locus [7]. Single nucleotide polymorphisms (SNPs) are common in the TNF-α gene, and a number of studies have been intensively focused on the promoter position −308 (G/A) and −238 (G/A) [8, 9]. However, because of the limitation of sample size and insufficient statistical power, there were inconsistent results among these studies. Recently, a meta-analysis, including eight studies, was performed for the evaluation of association between TNF-α promoter polymorphisms and AS [10]. However, new studies about the role of TNF-α promoter polymorphisms in AS have been published and provided new evidence that were not included in the previous meta-analysis [11–15]. Therefore, it is essential to recollect all the relevant studies and reevaluate the association of TNF-α promoter polymorphisms with AS. We performed this meta-analysis to update the results of the previous one.
Materials and methods
Search strategy and identification of relevant studies
We performed a literature research to identify publications, which are related to the association of TNF-α promoter polymorphisms with AS susceptibility, using the combinations of the following keywords: “tumor necrosis factor alpha,” “TNF-α,” “promoter,” “polymorphisms,” “ankylosing spondylitis,” “AS” without language restriction. Sources included Medline, Pubmed, Embase, and Web of Science for the period up to February, 2010 (last research: February 20, 2010). We also checked all the references of retrieved articles and contacted the investigators for additional data and explanations when key information relevant to the meta-analysis was missing.
Studies meeting the following criteria were included in the meta-analysis: (1) The type of study was case–control or cohort study. (2) The distributions of genotype or alleles on position −238 or −308 within the TNF-α promoter were provided for both AS patients and controls. (3) Basic characters of the participants including population source, age, gender, etc. were provided. The exclusion criteria are as follows: (1) The study was a case report, review, descriptive research, or meta-analysis. (2) Individuals in the studies were of family members. (3) Duplicate data were contained in the studies. (4) The participants involved in studies were fewer than ten.
Data extraction
Two of the authors extracted the information from all the studies meeting the inclusion criteria independently, and the following characters were collected: first author, country, year of publication, demographic characteristics, frequencies of genotypes and alleles on positions 238 and 308 within TNF-α promoter in cases and controls, and the method for testing genotype. Data were also extracted separately based on different ethnic groups and different HLA-B27 types of the individuals that were involved in studies. Disagreements were resolved by discussion.
Statistical analysis
The association between TNF-α polymorphisms and AS susceptibility was estimated by the odds ratios (OR) and 95% confidence interval (95%CI). For the 238 and 308 positions within the TNF-α promoter, two different ORs were calculated to evaluate the association of mutants with AS: (1) AA (homozygous mutants) + GA (heterozygous) vs GG (homozygous wild types); (2) AA vs GA+GG. We also evaluated the risk of A (minor allele) vs G (common allele) for the rare frequencies of A allele. In addition, separate analyses were performed in the population of different types of HLA-B27 (HLA-B27+ or HLA-B27−) and different ethnicities described in the previous study [10].
Both fixed effects model [16] and random effects model [17] were used to calculate the pool ORs. Random effects model was the better choice when heterogeneity was significant. We used Cochrane Q test to examine the heterogeneity among studies, which was considered significant if the p value was <0.1. I 2 statistic was also calculated to quantify heterogeneity between studies. In our meta-analysis, heterogeneity was defined “low” if the I 2 values were <25%, defined “moderate” if the I 2 values were between 25% and 75%, and values of larger than 75% represented “large” heterogeneity [18, 19]. The effect of publication bias was determined by funnel plot where the standard error of log(OR) of each study was plotted against its log(OR). Asymmetric plot indicated possible publication bias. Egger’s linear regression test, a linear regression approach to measure the Funnel plot asymmetry on natural logarithm scale of the OR, was used to evaluate the Funnel plot asymmetry. Statistical significance was considered when the p value of Egger’s test was <0.05 [20]. All of the calculations were conducted in the computer program Stata, version 10.0 (Stata Corporation, College Station, TX, USA), two-sided for all p values. A web-based program (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl) was used to test the departure of frequencies of TNF-α promoter polymorphisms (−238, −308) from expectation under Hardy–Weinberg equilibrium (HWE) in the control population.
Result
Characters of eligible studies
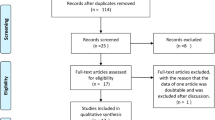
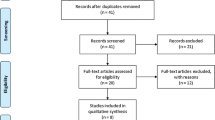
Based on the search criteria, 20 studies relevant to the role of TNF-α promoter polymorphisms in AS susceptibility were identified. Five of these articles were excluded: Four of these reports were review articles [21–24], and one report was a meta-analysis [10]. One study was excluded for the lack of control group [25]. Overall, 14 independent studies, including 1,766 AS patients and 2,114 healthy controls, were entered in the meta-analysis [11–15, 26–34]. The characteristics of each eligible study are shown in Table 1. The publishing year of the included studies ranged from 1994 to 2009. Among them, ten studies had been conducted in a European population [11, 13, 15, 26–29, 31–33], two studies were based on Asian population [12, 14], one study had been conducted in Mexico [33], and one study included different descendants [30]. Nine studies [11–15, 26–28, 32] used the “modified New York criteria” [35] as diagnostic criteria for AS, four studies [29, 31, 33, 34] used “New York criteria” [36] for the diagnosis of AS, and one study did not provide the exact clinical diagnostic criteria for AS patients [30]. Twelve studies provided information on the association between TNF-α promoter −238G/A polymorphic and AS susceptibility [11–15, 26–30, 32, 34], 13 studies were concerned with TNF-α promoter −308G/A polymorphism [11–15, 26–28, 30–34], two studies investigated the relation between TNF-α promoter −376 polymorphism and AS susceptibility [29, 32], one study for TNF-α promoter −244 polymorphism [26], and one for TNF-α −857 polymorphism [11]. Because of the insufficient sample populations available for TNF-α promoter −376, −244 polymorphism and TNF-α −857 polymorphism, we did not analyze these genetic polymorphisms. The study of Milicic et al. [32] sorted the data in English-European and Southern German-European; therefore, we considered these two groups independently in this meta-analysis. There were 11 studies that provided the number of different HLA-B27 (positive/negative) participants [11–13, 26–31, 33, 34], among which nine studies had the available data for meta-analysis [12, 26–31, 33, 34]. Because of the limited number of related studies and small sample size [27, 30], the association between TNF-α promoter polymorphism and AS susceptibility in HLA-B27− population was not estimated in our meta-analysis.
Statistics
The association of TNF-α promoter polymorphism with AS susceptibility in common population
−238A/G polymorphism and AS
As shown in Table 2, 11 and 6 studies were included in our meta-analysis for (AA+GA) vs GG and AA vs (GA+GG), respectively. For (AA+GA) vs GG, no statistical significance was observed (OR = 0.927, 95%CI = 0.647–1.326) where random model was used due to the heterogeneity between studies (Q = 22.45, p = 0.013, I 2 = 55.5%). For AA vs (GA+GG), statistical significance was observed (OR = 1.630, 95%CI = 1.116–2.381) in fixed model (Q = 2.071, p = 0.839, I 2 = 0.0%; Table 2). However, because of overweight of Nicknam’s study (weight = 87.93%), we recalculated the overall OR and 95%CI after excluding this study, and no association was found between AA genotype at position 238 of TNF-α promoter and AS (Fig. 1). Eleven studies, including 1,249 AS patients and 1,531 healthy controls, were included in the meta-analysis for the comparison of allele A and allele G in which the overall OR was 0.654 (95%CI = 0.385–1.111) in random model (Q = 37.14, p = 0.000, I 2 = 73.9%).
−308A/G polymorphism and AS
We did not find any statistical significance while evaluating the association between TNF-α promoter −308 polymorphism and AS susceptibility in the common population. The overall ORs for (AA+GA) vs GG, AA vs (GA+GG), and allele A vs allele G were 0.844 (95%CI 0.696–1.024), 1.013 (95%CI 0.716–1.433), and 0.759 (95%CI 0.563, 1.023), respectively. The fixed effect method was used in (AA+GA) vs GG and AA vs (GA+GG), and the random effect method was used in the comparison of allele A with allele G because of the significant heterogeneity between studies (Q = 32.542, p = 0.001, I 2 = 66.3%; Table 2).
The association between TNF-α promoter polymorphism and AS susceptibility in HLA-B27+ population
In the HLA-B27+ population, we did not find any statistical significance in the evaluation of association between TNF-α promoter −238 polymorphism and AS risk. The overall ORs of (AA+GA) vs GG and allele A vs allele G were 1.207 (95%CI 0.737–1.976) and 0.934 (95%CI 0.538–1.621) under the fixed effect model separately. For the TNF-α promoter −308 polymorphism in HLA-B27+ participants, results of meta-analysis indicated that there was statistical difference in the comparison of allele A with allele G (OR = 0.721, 95%CI = 0.522–0.995) in the fixed model (Q = 8.000, p = 0.238, I 2 = 25.0%). However, the overall OR for (AA+GA) vs GG did not show statistical significance (OR = 0.856, 95%CI = 0.613–1.195) in the fixed model (Q = 5.210, p = 0.390, I 2 = 4.1%; Table 2).
Hardy–Weinberg equilibrium
The results of the HWE test are shown in Table 3. For −308 polymorphisms, nine studies were available to perform the HWE test [11–14, 26, 27, 31, 32, 34], three of which were not in Hardy–Weinberg equilibrium [27, 31, 32]. For −238 polymorphisms, eight studies provided exact genotype distribution to test HWE [11–14, 26, 27, 32, 34], two of which were not in Hardy–Weinberg equilibrium [13, 32]. Sensitive analysis was carried out by limiting the studies conforming HWE, and all of the results were not materially changed except for 308 polymorphisms in HLA-B27+ population while regarding to allele A vs allele G. After adjustment, the previously decreased frequency of A allele was no longer statistically significant (OR = 1.150; 95%CI = 0.568–2.310; Fig. 2).
Publication bias
Publication bias of the literatures was accessed by Begg’s funnel plot and Egger’s test. For allele A vs allele G of the TNF-α promoter −238, Egger’s test showed that there was statistical significance for the evaluation of publication bias (p = 0.011). The shape of the funnel plot also showed some asymmetry (Fig. 3). These results suggested the evidence of potential publication bias. However, no significant publication bias was detected for the other comparisons (Table 2).
Discussion
Although there are a number of SNPS in the TNF-α promoter, the majority of studies have focused on two polymorphisms: −238G to A and −308G to A. In addition, the role of these genetic variants in AS susceptibility remained the subject of debate [11–15, 26–34]. A meta-analysis, which was conducted by Young et al. showed that there was no association of the TNF-α promoter 308 and 238 A/G polymorphisms with AS [10]. However, some studies after Young’s study have provided new evidence for the role of TNF-α promoter polymorphisms in AS susceptibility, with inconsistent results [11–15]. Our meta-analysis collected all the related studies up to date and reevaluated the association between TNF-α promoter polymorphisms (−238G/A and −308G/A) and AS.
Some studies have reported that the frequency of A allele at position 238 of TNF-α promoter decreased in AS patients compared with healthy controls, suggesting that A allele was a protective factor for AS [14, 15, 28]. It might be due to the increased level of TNF-α in the individuals carrying allele A at position 238, which could prevent the pathogenesis of AS infection with Klebsiella pneumoniae and other Gram-negative enterobacteria [37]. However, different from the aforementioned studies and the previous meta-analysis, we found that the AA genotype in position 238 was associated with AS susceptibility. The increased frequency of AA genotype in AS patients, independent of HLA-B27, suggested that people with AA genotype in position 238 of the TNF-α promoter were more susceptible to AS. Interestingly, we found that AA genotype, which was the homozygous mutant in the overall population, became the dominant type in Iranian in Nicknam’s study [13]. Maybe the different distribution of genotypes at position 238 in Iranian was due to the race difference and small simple size, but it was still difficult to explain the magnificent discrepancy. In addition, the weight of Nicknam’s study was over 80% in our meta-analysis, which made the meta-analysis unstable. After excluding this unstable factor, statistical significance was no longer observed (Fig. 1).
Although our meta-analysis indicated that there was no relationship between the TNF-α promoter 308 polymorphism and AS susceptibility in the overall population, the result in HLA-B27+ population showed that the frequency of allele A at position 308 modestly decreased in AS patients compared with healthy controls (Table 2). This result suggested that allele A at position 308 played a protective role in AS susceptibility regardless of the influence of HLA-B27, which was consistent with what had been described in McGarry’s and Hohler’s studies [28, 31]. The reasonable explanation for the inconsistent results between the overall population and HLA-B27+ population might be the TNF/HLA-B27 linkage disequilibrium due to their adjacent location [38]. In addition, this result was also in conflict with the previous study, which stated that no association was found between the B27-positive AS patients and the matched B27-positive controls for TNF-α 308 polymorphism. The reasons for inconsistent results may be the strengthened statistical power obtained by adding new evidence to the current meta-analysis or the limited genetic background in Young’s study(the majority of AS patients and healthy controls were Europeans) [10]. But after the sensitive analysis based on the result of the HWE test, the overall effect was no longer statistically significant (Fig. 2). According to the study of Trikalinos et al. [39], the meta-analysis of gene-related disease should be pronounced with extra caution when p values are not much smaller than 0.05 (p = 0.046 in our study) regarding the HWE-violating studies. Therefore, we still could not state that the frequency of A allele at position 308 decreased in AS patients compared with the healthy controls in HLA-b27+ population.
Between-study heterogeneity was observed for (AA+GA) vs GG, A vs G at position 238, and A vs G at position 308. But the reasons for the existence of heterogeneity were unclear. The source of heterogeneity could probably come from the study design, year of publication, and especially the ethics due to different genetic backgrounds, different linkage disequilibrium patterns, and clinical heterogeneity among different ethnic populations. It is now increasingly considered that the inconsistent results among studies are mainly caused by various ethnicities [13, 15], which had been classified as European and Latin American by Young et al. in the previous meta-analysis [10]. However, we found that the between-study heterogeneity in this meta-analysis could not be excluded by subgroup analysis using Young’s method (data not shown). Therefore, ethnicity may not play an important role in the generation of heterogeneity as considered previously, or some new classifications of the human race are needed to explain the heterogeneity instead of Young’s. Besides, there are many diagnostic criteria for AS, including the New York criteria, modified New York criteria, and Rome criteria. All of them are clinical criteria and could not provide an accurate initial diagnosis for AS due to some subjective items in these criteria, such as “limitation of the lumbar spine in three directions” and “limitation of chest expansion.” These items would be different for the same patient based on the definition of limitation and doctors’ experience [40]. So this could be another source of heterogeneity.
Some limitations should be acknowledged in our meta-analysis. First, the individuals with AA genotype, which is a homozygous mutant for both positions 238 and 308 in the TNF-α promoter, only took a small percentage of the overall population. Under the condition of small sample size, the rare frequency of AA genotype might not have enough statistical power to explore the real association between TNF-α polymorphisms and AS susceptibility, especially for HLA-B27+ population. Secondly, the gene distribution in AS patients and healthy controls in Nicknam’s study were significantly different from other studies included in the meta-analysis, and there was no reasonable explanation for it. Thirdly, only published articles were included in our meta-analysis. Therefore, publication bias may have occurred. Lastly, only two studies investigated the role of TNF-α polymorphisms in AS in the HLA-B27-negative population [27, 30]. Because of the limited information, we were not able to evaluate the relationship of them in the HLA-B27-negative population.
Taken together, our meta-analysis concluded that no association was found between TNF-α promoter 238/308 polymorphisms and ankylosing spondylitis susceptibility in both the overall and HLA-B27+ population.
References
Brewerton DA, Hart FD, Nicholls A, Caffrey M, James DC, Sturrock RD (1973) Ankylosing spondylitis and HL-A 27. Lancet 1:904–907
Khan MA, Ball EJ (2002) Genetic aspects of ankylosing spondylitis. Best Pract Res Clin Rheumatol 16:675–690
Rubin LA, Amos CI, Wade JA et al (1994) Investigating the genetic basis for ankylosing spondylitis. Linkage studies with the major histocompatibility complex region. Arthritis Rheum 37:1212–1220
van der Linden SM, Valkenburg HA, de Jongh BM, Cats A (1984) The risk of developing ankylosing spondylitis in HLA-B27 positive individuals. A comparison of relatives of spondylitis patients with the general population. Arthritis Rheum 27:241–249
Benjamin R, Parham P (1990) Guilt by association: HLA-B27 and ankylosing spondylitis. Immunol Today 11:137–142
Tracey KJ, Cerami A (1993) Tumor necrosis factor: an updated review of its biology. Crit Care Med 21:S415–S422
Glossop JR, Nixon NB, Dawes PT, Hassell AB, Mattey DL (2003) No association of polymorphisms in the tumor necrosis factor receptor I and receptor II genes with disease severity in rheumatoid arthritis. J Rheumatol 30:1406–1409
Wilson AG, de Vries N, Pociot F, di Giovine FS, van der Putte LB, Duff GW (1993) An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA a1, B8, and DR3 alleles. J Exp Med 177:557–560
D'Alfonso S, Richiardi PM (1994) A polymorphic variation in a putative regulation box of the TNFa promoter region. Immunogenetics 39:150–154
Lee YH, Song GG (2009) Lack of association of TNF-alpha promoter polymorphisms with ankylosing spondylitis: a meta-analysis. Rheumatology (Oxford) 48:1359–1362
Chatzikyriakidou A, Georgiou I, Voulgari PV, Drosos AA (2009) The role of tumor necrosis factor (TNF)-alpha and TNF receptor polymorphisms in susceptibility to ankylosing spondylitis. Clin Exp Rheumatol 27:645–648
Lu MC, Yang KL, Tung CH et al (2008) Higher LPS-stimulated TNF-alpha mRNA levels in peripheral blood mononuclear cells from Chinese ankylosing spondylitis patients with −308G/A polymorphism in promoter region of tumor necrosis factor: association with distinct A33/B58/Cw10 haplotypes. Rheumatol Int 29:189–195
Nicknam MH, Mahmoudi M, Amirzargar AA, Jamshidi AR, Rezaei N, Nikbin B (2009) HLA-B27 subtypes and tumor necrosis factor alpha promoter region polymorphism in Iranian patients with ankylosing spondylitis. Eur Cytokine Netw 20:17–20
Shiau MY, Lo MK, Chang CP, Yang TP, Ho KT, Chang YH (2007) Association of tumour necrosis factor alpha promoter polymorphisms with ankylosing spondylitis in Taiwan. Ann Rheum Dis 66:562–563
Sousa E, Caetano-Lopes J, Pinto P et al (2009) Ankylosing spondylitis susceptibility and severity—contribution of TNF gene promoter polymorphisms at positions −238 and −308. Ann N Y Acad Sci 1173:581–588
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21:1539–1558
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560
Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Aguillon JC, Cruzat A, Aravena O, Salazar L, Llanos C, Cuchacovich M (2006) Could single-nucleotide polymorphisms (SNPs) affecting the tumour necrosis factor promoter be considered as part of rheumatoid arthritis evolution? Immunobiology 211:75–84
De Vos M (2004) Review article: joint involvement in inflammatory bowel disease. Aliment Pharmacol Ther 20(Suppl 4):36–42
Rudwaleit M, Hohler T (2001) Cytokine gene polymorphisms relevant for the spondyloarthropathies. Curr Opin Rheumatol 13:250–254
Field M (2001) Tumour necrosis factor polymorphisms in rheumatic diseases. QJM 94:237–246
Seitz M, Wirthmuller U, Moller B, Villiger PM (2007) The −308 tumour necrosis factor-alpha gene polymorphism predicts therapeutic response to TNFalpha-blockers in rheumatoid arthritis and spondyloarthritis patients. Rheumatology (Oxford) 46:93–96
Fraile A, Nieto A, Beraun Y, Vinasco J, Mataran L, Martin J (1998) Tumor necrosis factor gene polymorphisms in ankylosing spondylitis. Tissue Antigens 51:386–390
Gonzalez S, Torre-Alonso JC, Martinez-Borra J et al (2001) TNF-238a promoter polymorphism contributes to susceptibility to ankylosing spondylitis in HLA-B27 negative patients. J Rheumatol 28:1288–1293
Hohler T, Schaper T, Schneider PM, Meyer ZBK, Marker-Hermann E (1998) Association of different tumor necrosis factor alpha promoter allele frequencies with ankylosing spondylitis in HLA-B27 positive individuals. Arthritis Rheum 41:1489–1492
Kaijzel EL, Brinkman BM, van Krugten MV et al (1999) Polymorphism within the tumor necrosis factor alpha (TNF) promoter region in patients with ankylosing spondylitis. Hum Immunol 60:140–144
Martinez-Borra J, Gonzalez S, Lopez-Vazquez A et al (2000) HLA-B27 alone rather than B27-related class I haplotypes contributes to ankylosing spondylitis susceptibility. Hum Immunol 61:131–139
McGarry F, Walker R, Sturrock R, Field M (1999) The −308.1 polymorphism in the promoter region of the tumor necrosis factor gene is associated with ankylosing spondylitis independent of HLA-B27. J Rheumatol 26:1110–1116
Milicic A, Lindheimer F, Laval S et al (2000) Interethnic studies of tnf polymorphisms confirm the likely presence of a second mhc susceptibility locus in ankylosing spondylitis. Genes Immun 1:418–422
Vargas-Alarcon G, Casasola-Vargas J, Rodriguez-Perez JM et al (2006) Tumor necrosis factor-alpha promoter polymorphisms in Mexican patients with spondyloarthritis. Hum Immunol 67:826–832
Verjans GM, Brinkman BM, Van Doornik CE, Kijlstra A, Verweij CL (1994) Polymorphism of tumour necrosis factor-alpha (TNF-alpha) at position −308 in relation to ankylosing spondylitis. Clin Exp Immunol 97:45–47
van der Linden S, Valkenburg HA, Cats A (1984) Evaluation of diagnostic criteria for ankylosing spondylitis, A proposal for modification of the New York criteria. Arthritis Rheum 27:361–368
Moll JM, Wright V (1973) New York clinical criteria for ankylosing spondylitis. A statistical evaluation. Ann Rheum Dis 32:354–363
Reveille JD (1993) The interplay of nature versus nurture in predisposition to the rheumatic diseases. Rheum Dis Clin North Am 19:15–27
Pei J, Choo SY, Spies T, Strominger JL, Hansen JA (1991) Association of four HLA class III region genomic markers with HLA haplotypes. Tissue Antigens 37:191–196
Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP (2006) Impact of violations and deviations in Hardy–Weinberg equilibrium on postulated gene–disease associations. Am J Epidemiol 163:300–309
Goie TH, Steven MM, van der Linden SM, Cats A (1985) Evaluation of diagnostic criteria for ankylosing spondylitis: a comparison of the Rome, New York and modified New York criteria in patients with a positive clinical history screening test for ankylosing spondylitis. Br J Rheumatol 24:242–249
Disclosures
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, B., Wang, P. & Li, H. The association between TNF-α promoter polymorphisms and ankylosing spondylitis: a meta-analysis. Clin Rheumatol 29, 983–990 (2010). https://doi.org/10.1007/s10067-010-1499-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-010-1499-y